Amphiphysin2 recruits clathrin to membranes
This material is supplemental to Peter et al., "BAR Domains as Sensors of Membrane Curvature: The Amphiphysin BAR Structure" Science Express Nov 26 (abstract); print version Jan 23, 2004.
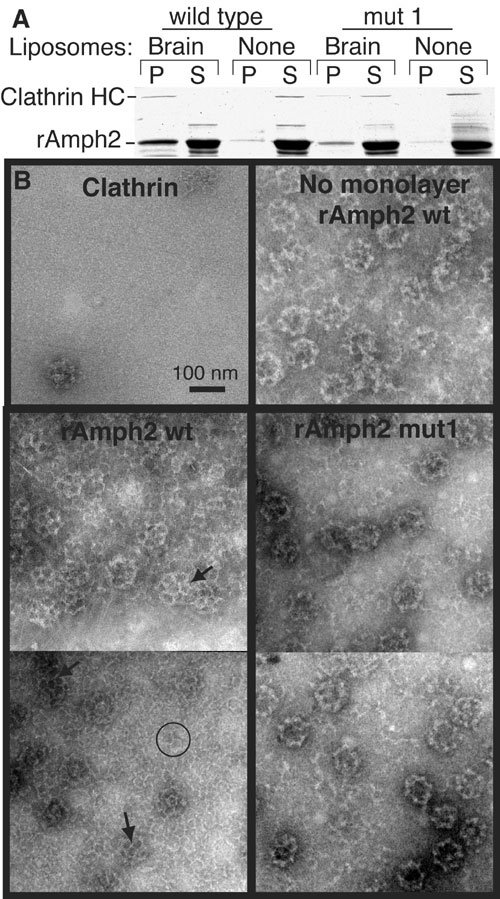
Fig. S4. Recruitment and polymerization of clathrin by amphiphysin
(A) Rat amphiphysin2 (rAmph2) was incubated with rat brain clathrin in the presence or absence of brain liposomes before sedimentation at 140000xg for 15min. In the Coomassie-stained gel, the first four lanes show the results obtained with wild-type protein and the last four lanes show the results obtained with the BAR mutant. Proteins in the pellet (P) or supernatant (S) were separated by SDS-PAGE. rAmph2 mut1 has the mutations K164E+K166E and was much less effective at recruiting clathrin to the membrane. Under these conditions no clathrin sedimented in the absence of other proteins. (B) Lipid monolayer assay demonstrating the ability of rAmph2 to recruit and polymerize clathrin. In the absence of amphiphysin, very little clathrin was on the monolayer, although occasionally a clathrin cage was seen (top left panel). When wild type rAmph2 was incubated with clathrin in the absence of lipid, the rAmph2 induced extensive polymerization of the clathrin into cages, which stuck to the grid (top right panel). However, in the presence of brain lipids, rAmph2 recruited clathrin and induced the formation of extensive lattices on the monolayers (lower left panel). The monolayers now had a high background of clathrin, and some individual clathrin triskelia were discerned (circle). Furthermore much of the clathrin was polymerized into lattice structures on the monolayer which were slightly invaginated given the presence of pentagons (arrows). In contrast the mut1 rAmph2 monolayers showed a lower background of triskelia, and now all of the polymerized clathrin was in the form of cages. Cages sitting on the surface of the monolayer can be distinguished from lattices by (a) their more defined edges, (b) their higher electron density owing to accumulated stain inside the cage, and (c) less definition of their triskelia, together resulting in a ringlike appearance. Thus mut1 rAmph2 still polymerized clathrin in solution, which is expected as the dimerization interface and clathrin binding sites on amphiphysin were unchanged. However, mutations in the BAR seriously inhibited the ability of amphiphysin to form ordered clathrin structures on the membrane.
(click on image to see a higher resolution version (824 kb) in a new window)