Research
in the Passmore Lab
Research
in the Passmore Lab

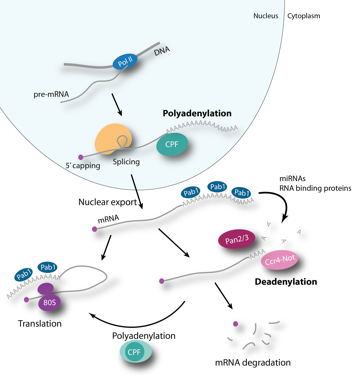
The Passmore laboratory uses a integrated approach to study the structure and function of multi-protein complexes involved in mRNA polyadenylation and deadenylation. Eukaryotic genes are normally transcribed as pre-mRNAs that must be processed before they are exported from the nucleus and translated into proteins. The 3´-end of the pre-mRNA is cleaved and a poly(A) tail is added. Several protein factors are required to
co-ordinate the 3´-end processing reactions, and to facilitate communication with the transcription and splicing machinery. This includes a large multi-protein complex called Cleavage and Polyadenylation Factor (CPF/CPSF).
Macromolecular machines that regulate mRNA 3´ ends
Macromolecular machines involved in DNA repair
We are also interested in protein complexes involved repair of damaged DNA. We are studying the Fanconi Anaemia core complex, a multi-protein E3 monoubiquitin ligase involved in the repair of DNA crosslinks and we have discovered that FANCD2-FANCI is a DNA clamp.


The poly(A) tail is required for export of the mRNA into the cytoplasm. It also enhances mRNA stability and stimulates translation, and its length can be regulated to influence these functions. For example, shortening of the poly(A) tail can decrease the efficiency of translation to control gene expression in some cases, and is the first step in mRNA turnover. The main deadenylase activities are found within the evolutionarily conserved, 1 MDa Ccr4-Not complex and the Pan2–Pan3 complex.
By understanding the structure and mechanisms of these complexes, we will gain insight into the regulation of mRNA polyadenylation and gene expression.
Cryo-EM structure of the polyadenylation module of the CPF
(Casañal, Kumar et al 2017)
Cryo-EM structure of the FA core complex
(Shakeel, Rajendra et al 2019)
Image by Jeroen Claus (Phospho Biomedical Animation)

Passmore Lab, MRC Laboratory of Molecular Biology, Cambridge UK