Coot 1.1 - Tutorial 1
Lucrezia Catapano & Paul Emsley - March 2025
Let’s do the Coot Tutorial using modern methods!
Before we start, familiarise yourself with the basic navigation and keyboard shortcuts in Coot 1.1:
Basic Navigation - Mouse & Keyboard Shortcuts.
1. Load the data
Load the tutorial:
- Calculate → Load Tutorial Model and Data
The map is quite nice looking - the resolution of the data is 1.8 Å.
Let’s turn on Updating Maps (so that the maps track the current model):
- Calculate → Updating Maps
- Check the Auto-Update button
- OK
2. Check the problems
Let’s view the structure anomalies:
- Validate → Overlaps, Peptides, CBeta Ramas & Rotas
A dialog is presented with Outliers listed by chain-id and residue number
Note: This structure is old, at which time the criteria for chiral volumes outliers was less strict than today standards. So, let’s ignore the chiral volume outliers for now.
Click on the first button (for A 2). Take a look at what is interacting with this VAL. See that it’s actually the PHE at A 89 that’s wrong. Recentre on that residue (middle-mouse click on the CB).
3. Fix and Refine
- Click on Refine - RSR Sphere on the vertical toolbar
- or Shift R (on the keyboard)
The figure shows the new types of geometry validation markups 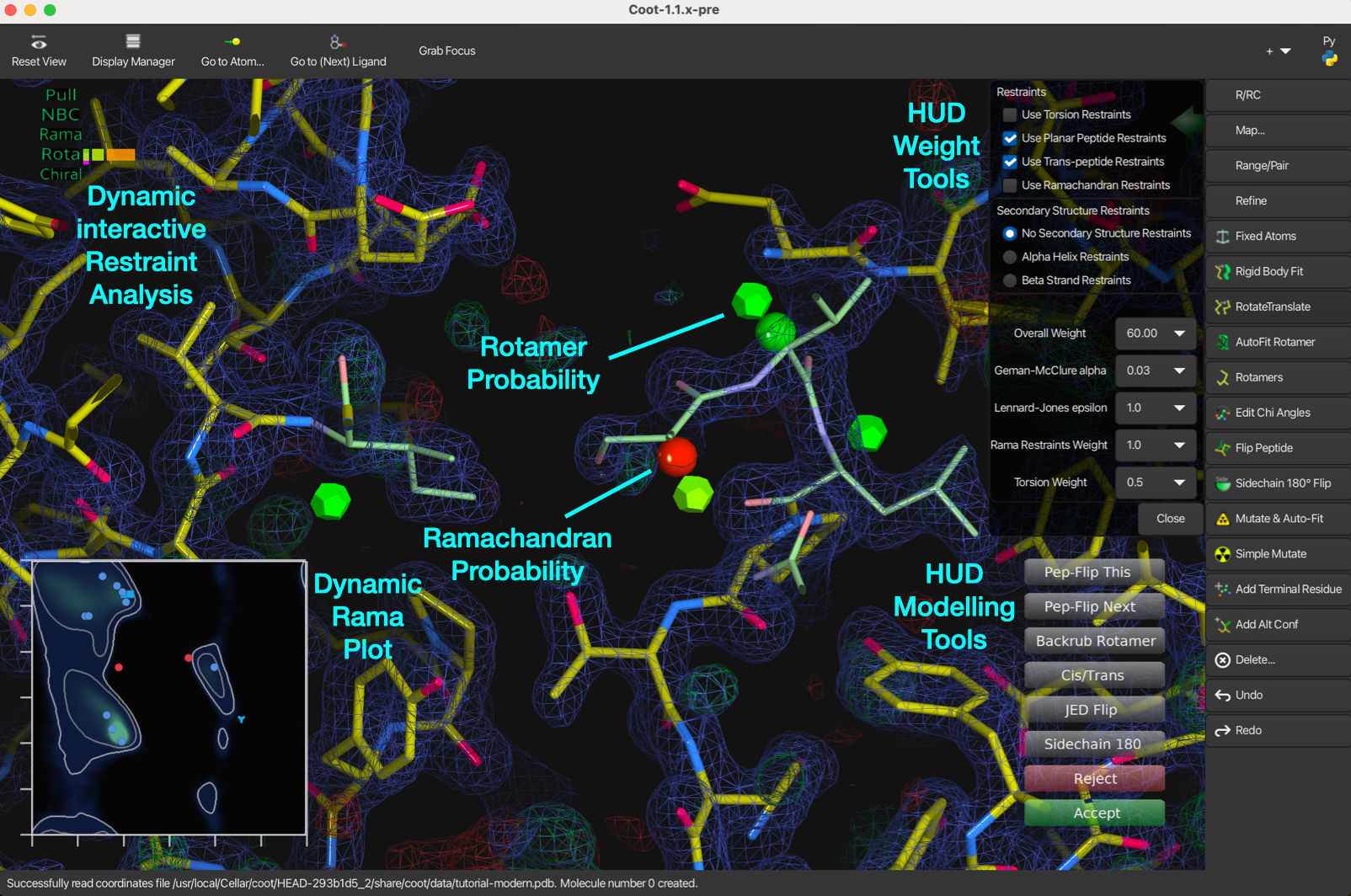
Fix the rotamer:
- Click on Backrub Rotamer
- or E (on the keyboard)
- Accept button or Enter/Return key
The sidechain will jump into the density, the maps will update and the difference maps will disappear in this region.
A new overlay with Coot Points, and R factors, will appear. You will collect Coot points as the model improves.
Note: The R factors are calculated by a clipper function, not REFMAC5.
Next problem in the dialog is at A 32. The density here is not very good and there may be a number of different conformers/rotamers. Let’s, for now, just choose one of them that has higher probability than the current model.
- E (on the keyboard)
- Enter/Return key
Notice that the number of Outliers/Problems has decreased.
Now look at the rest of the labels in that dialog. Many of them are around the same residues, 40, 41 and 42.
- Shift R (for sphere refine)
Let’s look at the Ramachandran Plot for this chain.
Let’s click on the first red spot on the left.
In the refinement dialog:
- Pep-Flip this
- Accept
The Rama balls go green. The 2 outliers (red spots) in the Ramachandran plot are now gone.
Let’s fix the other rotamer outliers, ignoring the chiral volumes outliers.
Finally, click on Refine - RSR Chain to refine the entire chain. If you reopen the validation dialog, you will see that all the chiral volume issues are resolved.
Let’s have a look at some missing atoms:
- Ctrl G and type A72.
Here we have missing main-chain atoms and missing side-chain atoms
- Delete the residue using Ctrl D
- Add in a replacement ALA using Y
- Shift R to refine
- Accept
- Mutate & Auto-fit on the vertical toolbar
- click on an atom in the new ALA - change it to a “CYS”
- Add Alt Conf on the vertical toolbar.. and split the whole residue
- Rotamer number 3 is the correct one
- Shift R to refine
4. Unmodelled blobs
Let’s find parts of the map that are as yet not modelled, i.e. find unmodelled blobs:
- Validate → Unmodelled blobs
- Defaults are fine
- Find Blobs
Blobs are sorted by size.
- Click on the button Blob 1
What are we looking at?
Blob 4 (if present in the dialog) and Blob 3 are the same thing. Let’s do Blob 3 first because the density is better. Notice the shape of the blobs and the environment. The precipitant for this structure was ammonium sulfate. So let’s add a SO4 here:
- Modelling → Add Other Solvent Molecules
- SO4: SULPHATE ION
Coot adds a sulfate ion using “Jiggle Fit”
Now you can do the same thing for Blob 4 if you wish.
5. Fitting the ligand
Blob 2 is the ligand. Let’s build a ligand into that:
- File → Get Monomer
- 3GP (this is 3’-guanosine monophosphate)
- OK
Check the chemistry (yes the phosphate is on the 3’ end).
Undisplay the new 3GP using the Display Manager
Modelling → Find Ligands :
In the following dialog, check the button for the 3GP_from_dict ligand (no need to make it flexible).
In “Where to Search?” choose “Right Here”.
Go to the Display Manager, and there will be Fitted-ligand in the molecule window.
When the ligand is corrected placed, let’s merge it into the main molecule:
- Edit → Merge Molecules
- Choose the fitted ligand and add it to molecule 0 (the protein)
6 . Missing Residues
Blob 1 is a piece of missing protein.
- Draw → Molecule → Sequence View and pull across to the protein model.
You can see on the right hand side of the sequence view that the A chain is missing the residues “QTC”
- Middle-mouse click the “C” atom of residue A 93.
- Calculate → Fit Loop → Fit Loop by Ramachandran Search…
- Choose Residue numbers 94 and 96 in the A chain
- Add the sequence: QTC
- Fit Loop
Coot builds the loop!
Is there still work to be done?
Maybe. You can use Ctrl Shift Arrow keys to reorient the CYS A 96 if needed (Use Ctrl Arrow keys to rotate the residue (about an axis that’s into the screen))
- Modelling → Add OXT to Residue…
- Add it