Regulation of phosphoinositide 3-kinases
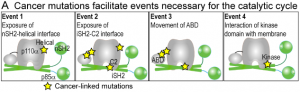
The PI3Ks are activated downstream of receptor tyrosine kinases (RTKs), G-protein coupled receptors (GPCRs) and members of the Ras-superfamily G-proteins. Because these regulatory interactions are specific to individual isoforms, it may be that they represent a useful target for isoform-specific therapeutics. Although we are determining the structures of complexes of these regulators with PI3Ks using X-ray crystallography, understanding the mechanism of the regulation is intricately tied in with the interactions that these complexes make with membranes and the influence of the regulators on the dynamics of the PI3Ks. In order to analyse the structural mechanisms of activation, hydrogen-deuterium exchange mass spectrometry and other biophysical approaches has been an invaluable component of our work.
Autophagy
One major direction of the group is the structures and functions of protein complexes involved in autophagy. Our work begain with the structure of the core of one of these complexes, the class III PI3K, Vps34.
Autophagy is an ancient process that is present in all eukaryotic cells and it intimately linked with nutrient sensing. The mammalian signalling component of the PI3K pathway is a more recent addition, but it shares intricate connections with autophagy and nutrient sensing. Autophagy acts as a survival mechanism during starvation, and it can act as a quality-control system to remove unneeded organelles or protein aggregates. There is a close connection between autophagy and life span extension, and the process is required for preventing age-related brain degeneration.
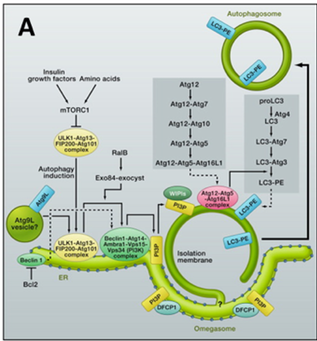
Autophagy is associated with cancer, but it is complex and likely to be dependent on the tumour and the stage of its growth. During starvation, the protein kinase mTOR is inactivated. This triggers formation of double membrane vesicles termed autophagosomes that engulf cytosolic material and deliver it to lysosomes for degradation. A large number of genes are involved in autophagy, but work is underway to understand how these genes become engaged in autophagy and how the process is regulated both temporally and spatially (Fig A, From Mizushima and Kimatsu (2011) Cell 147, 729).
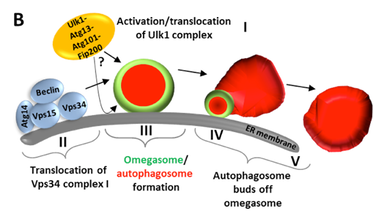
Ttwo protein complexes regulate the iinitial stages of autophagy. One complex that is sometimes callet the “initiation” complex consists of the protein kinase ULK1 and its associated subunits, Atg13, Atg101 and FIP200. The second complex contains the class III PI3K, Vps34 and three assoxiated proteins, Vps15, beclin and Atg14. This second complex, sometimes called the “nucleation complex” can give rise to “omegasomes”, which are membrane extensions of the endoplasmic reticulum (ER) enriched in PI3P. Within these omegasomes, the first autophagosomes appear and then bud off (Fig B, courtesy of NIck Ktistakis). The initiation complex targets autophagosomal membranes, but its relation to the nucleation complex and the lipid kiase activity of the nucleation complex is still under investigation.