Local symmetry
TODO Shaoda
Introduction
Helical processing in RELION closely follows the procedures normally applied for single-particle analysis. Therefore, if you are new to RELION, it is highly recommended that you first go through the tutorial. A paper about the details of helical processing in RELION is now available on JSB ScienceDirect. Below is an overview of specific parameters and issues with helical processing in RELION.
(Please note that GPU acceleration is applicable in 3D reconstructions of 2D helical segments. While 3D sub-tomogram averaging with helical parameters is currently under development.)
Description of parameters
Helical symmetry
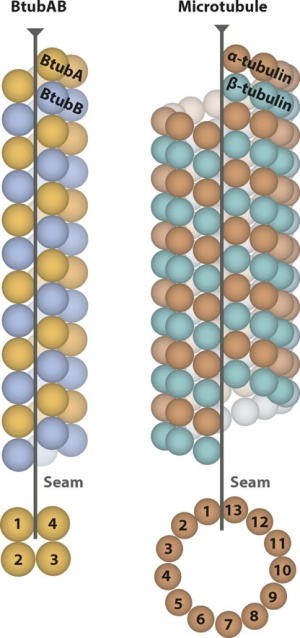
$$$ Example: 3-start 13-microtubule with seam $$$
Local symmetry STAR file (12 operators): ftp://ftp.mrc-lmb.cam.ac.uk/pub/she/localsym/localsym-3s13m.star
The only mask (binary, soft-edged, pixel size = 1.38 Angtroms): ftp://ftp.mrc-lmb.cam.ac.uk/pub/she/localsym/mask-7.mrc
The referential density map (pixel size = 1.38 Angstroms): ftp://ftp.mrc-lmb.cam.ac.uk/pub/she/localsym/ref-fil30.mrc
Although the map here has been low-pass filtered to 30 Angstroms (unpublished results from another group), it is recommended to use the best map you have to set up the local symmetry. Always display the referential map and the mask(s) simultaneously in UCSF Chimera for a check after you have constructed the mask(s).
Note: Assume that as seen in the top-view/bottom-view, the proto-filaments are numbered as #1~#13 clockwise/counter-clockwise. The proto-filaments closest to the seam are #1 and #13 while the one opposite to the seam is numbered as #7. The only mask covers the tubulins (central ~50% of the box) along the proto-filament #7. The helical symmetry is defined along each proto-filament: twist = +0.1 degrees, rise = 83.1 Angstroms (with each pair of ab-tubulins treated as a subunit, or the building block of each proto-filament). In the local symmetry STAR file, the rot (or psi) angles are N*360/13 = N*27.69 degrees and the z translations are N*83.1*1.5/13 = N*9.59 Angstroms (N=-6~+6, without 0). All the other angles and translations are set to 0. The 12 operators (entries, each consisting of a rot angle and a z translation here) show how to move the whole proto-filament #7 onto any of the other 12 by a rotation around the helical axis and a translation along the axis.
It doesn't matter if you don't have a microtubule with seam as the starting reference. Given a referential 3-start 13-microtubule WITHOUT seam, you can just treat any proto-filament as #7 and the above mask and operators remain the same. The seam should become discernible after 3D auto-refinement if the local symmetry settings are correct.
Enable the local symmetry option for such structure: Copy the referential density map, the mask and the local symmetry STAR file to the project directory. Add '--local_symmetry localsym-3s13m.star --tau_fudge 13' as the additional option in 3D auto-refinement. We are still testing how '--tau_fudge' influences the 3D reconstructions with local symmetry. Unreasonably large values lead to over-fitting but the subsequent post-processing step will always give you a fair resolution estimate.