Pixel size issues: Difference between revisions
(RELION 3.1) |
m (absolute pixel size) |
||
| Line 1: | Line 1: | ||
== How can I merge datasets with different pixel sizes? | = What should I do if the pixel size turned out to be wrong? = | ||
If the error is small (say 1-2 %) and the resolution is not very high (3 Å), you can specify the correct pixel size in the PostProcess job. This scales the resolution in the FSC curve. You '''should not''' edit your STAR files because your current defocus values are fitted against the initial, slightly wrong pixel size. Also, you should '''not''' use "Manually set pixel size" in the Extraction job. It will make metadata inconsistent and break Bayesian Polishing. | |||
When the error is large, the presence of spherical aberration invalidates this approach. Continue reading. | |||
= Cs and the error in the pixel size = | |||
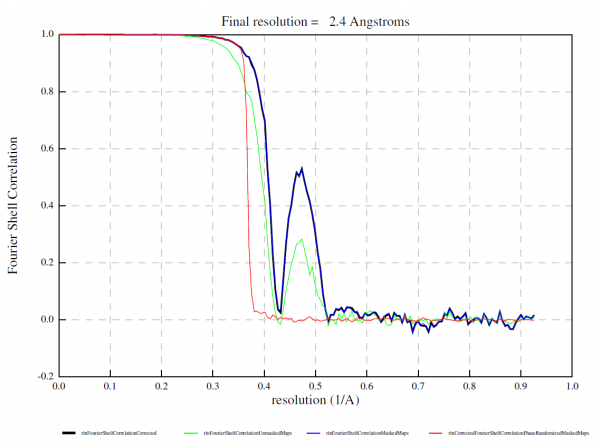
Recall that the phase shift due to defocus is proportional to the square of the wave number (i.e. inverse resolution), while that due to spherical aberration is proportional to the forth power of the wave number. At lower resolutions, the defocus term dominates and errors in the pixel size (i.e. errors in the wave number) can be absorbed into the defocus value fitted at the nominal pixel size. At higher resolution, however, the Cs term becomes significant. Since Cs is not fitted but given at the correct pixel size, the error persists. As the two terms have the opposite sign, the errors sometimes cancel out at certain resolution shells, leading to a strange bump in the FSC curve. See an example below contributed from a user. Here the pixel size was off by about 6 % (truth: 0.51 Å / px, nominal: 0.54 Å / px). | |||
[[File:FSC-bump.png|600px]] | |||
Below is a theoretical consideration. Let's consider a CTF at defocus 5000 Å, Cs 2.7 mm at 0.51 Å / px. This is shown in orange. | |||
If one thought the pixel size is 0.54 Å / px, the calculated CTF (blue) became quite off even at 5 Å (0.2). | |||
[[File:pixel-error-1.png]] | |||
However, the defocus is fitted (by CtfFind, followed by CtfRefine) at 0.54 Å / px, the nominal pixel size. The defocus became 5526 Å, absorbing the error in the pixel size. This is shown in blue. The fit is almost perfect up to about 3.3 Å. In this region, you can update the pixel size in PostProcess. Beyond this point, the error from the Cs term starts to appear and the two curves go out of phase. This is why the FSC drops to zero. However, the two curves came into phase again at about 1.8 Å (0.55)! This is why the FSC goes up again. | |||
[[File:pixel-error-2.png]] | |||
If you refine Cs and defocus simultaneously, the error in the pixel size is completely absorbed and the fit becomes perfect. Notice that the refined Cs is 3.39, which is 2.7 * (0.54 / 0.51)^4. Also note that the refined defocus 5606 Å is 5000 * (0.54 / 0.51)^2. | |||
[[File:pixel-error-3.png]] | |||
In practice, one just needs to run <code>CtfRefine</code> twice: first with <code>Estimate 4th order aberrations</code> to refine Cs, followed by another run for defocus. Note that <code>rlnSphericalAberration</code> remains the same. The error in Cs is expressed in <code>rlnEvenZernike</code>. You '''should never''' edit the pixel size in the STAR file! | |||
Now everything is consistent at the nominal pixel size of 0.54 Å/px. In PostProcess, one should specify 0.51 Å/px to re-scale the resolution and the header of the output map. | |||
= How can I merge datasets with different pixel sizes? = | |||
For RELION 3.0, please see an excellent explanation posted to CCPEM by Max Wilkinson: https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=CCPEM;fd0e7fab.1810. | For RELION 3.0, please see an excellent explanation posted to CCPEM by Max Wilkinson: https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=CCPEM;fd0e7fab.1810. | ||
| Line 58: | Line 87: | ||
Refine this combined dataset. For a reference and mask, you can use the pixel size and box size of either dataset 1 or 2. The output pixel size and the box size will be the same as the input reference map. After refinement, run <code>CtfRefine</code> with <code>Estimate anisotropic magnification: Yes</code>. This will refine the ''relative'' pixel size difference between two datasets. In the above example, the nominal difference is 10 %, but it might be actually 9.4 %, for example. Then run Refine3D again. The ''absolute'' pixel size of the output can drift a bit. It needs to be calibrated against atomic models. | Refine this combined dataset. For a reference and mask, you can use the pixel size and box size of either dataset 1 or 2. The output pixel size and the box size will be the same as the input reference map. After refinement, run <code>CtfRefine</code> with <code>Estimate anisotropic magnification: Yes</code>. This will refine the ''relative'' pixel size difference between two datasets. In the above example, the nominal difference is 10 %, but it might be actually 9.4 %, for example. Then run Refine3D again. The ''absolute'' pixel size of the output can drift a bit. It needs to be calibrated against atomic models. | ||
= | = How can I estimate the absolute pixel size? = | ||
Experimentally and ideally, one can use diffraction from calibration standards. | |||
Computationally, one can compare the map and a refined atomic model. However, if the model has been refined against the same map, the model might have been biased towards the map. Also note that bond and angle RMSDs are not always reliable when restraints are too strong. | |||
Revision as of 15:03, 10 October 2019
What should I do if the pixel size turned out to be wrong?
If the error is small (say 1-2 %) and the resolution is not very high (3 Å), you can specify the correct pixel size in the PostProcess job. This scales the resolution in the FSC curve. You should not edit your STAR files because your current defocus values are fitted against the initial, slightly wrong pixel size. Also, you should not use "Manually set pixel size" in the Extraction job. It will make metadata inconsistent and break Bayesian Polishing.
When the error is large, the presence of spherical aberration invalidates this approach. Continue reading.
Cs and the error in the pixel size
Recall that the phase shift due to defocus is proportional to the square of the wave number (i.e. inverse resolution), while that due to spherical aberration is proportional to the forth power of the wave number. At lower resolutions, the defocus term dominates and errors in the pixel size (i.e. errors in the wave number) can be absorbed into the defocus value fitted at the nominal pixel size. At higher resolution, however, the Cs term becomes significant. Since Cs is not fitted but given at the correct pixel size, the error persists. As the two terms have the opposite sign, the errors sometimes cancel out at certain resolution shells, leading to a strange bump in the FSC curve. See an example below contributed from a user. Here the pixel size was off by about 6 % (truth: 0.51 Å / px, nominal: 0.54 Å / px).
Below is a theoretical consideration. Let's consider a CTF at defocus 5000 Å, Cs 2.7 mm at 0.51 Å / px. This is shown in orange. If one thought the pixel size is 0.54 Å / px, the calculated CTF (blue) became quite off even at 5 Å (0.2).
However, the defocus is fitted (by CtfFind, followed by CtfRefine) at 0.54 Å / px, the nominal pixel size. The defocus became 5526 Å, absorbing the error in the pixel size. This is shown in blue. The fit is almost perfect up to about 3.3 Å. In this region, you can update the pixel size in PostProcess. Beyond this point, the error from the Cs term starts to appear and the two curves go out of phase. This is why the FSC drops to zero. However, the two curves came into phase again at about 1.8 Å (0.55)! This is why the FSC goes up again.
If you refine Cs and defocus simultaneously, the error in the pixel size is completely absorbed and the fit becomes perfect. Notice that the refined Cs is 3.39, which is 2.7 * (0.54 / 0.51)^4. Also note that the refined defocus 5606 Å is 5000 * (0.54 / 0.51)^2.
In practice, one just needs to run CtfRefine twice: first with Estimate 4th order aberrations to refine Cs, followed by another run for defocus. Note that rlnSphericalAberration remains the same. The error in Cs is expressed in rlnEvenZernike. You should never edit the pixel size in the STAR file!
Now everything is consistent at the nominal pixel size of 0.54 Å/px. In PostProcess, one should specify 0.51 Å/px to re-scale the resolution and the header of the output map.
How can I merge datasets with different pixel sizes?
For RELION 3.0, please see an excellent explanation posted to CCPEM by Max Wilkinson: https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=CCPEM;fd0e7fab.1810.
From RELION 3.1, you can refine particles with different pixel sizes and/or box sizes. Suppose you want to join two particle STAR files. First, make sure they have different rlnOpticsGroupName. For example:
Dataset1.star:
data_optics
loop_
_rlnOpticsGroup #1
_rlnOpticsGroupName #2
_rlnAmplitudeContrast #3
_rlnSphericalAberration #4
_rlnVoltage #5
_rlnImagePixelSize #6
_rlnMicrographOriginalPixelSize #7
_rlnImageSize #8
_rlnImageDimensionality #9
1 dataset1 0.100000 2.700000 300.000000 1.000000 1.000000 140 2
Dataset2.star:
data_optics
loop_
_rlnOpticsGroup #1
_rlnOpticsGroupName #2
_rlnAmplitudeContrast #3
_rlnSphericalAberration #4
_rlnVoltage #5
_rlnImagePixelSize #6
_rlnMicrographOriginalPixelSize #7
_rlnImageSize #8
_rlnImageDimensionality #9
1 dataset2 0.100000 2.700000 300.000000 1.100000 1.100000 128 2
Then use JoinStar. The result should look like:
data_optics
loop_
_rlnOpticsGroup #1
_rlnOpticsGroupName #2
_rlnAmplitudeContrast #3
_rlnSphericalAberration #4
_rlnVoltage #5
_rlnImagePixelSize #6
_rlnMicrographOriginalPixelSize #7
_rlnImageSize #8
_rlnImageDimensionality #9
1 dataset1 0.100000 2.700000 300.000000 1.000000 1.000000 140 2
2 dataset2 0.100000 2.700000 300.000000 1.100000 1.100000 128 2
Note that the dataset2's rlnOpticsGroup has been re-numbered to 2.
Refine this combined dataset. For a reference and mask, you can use the pixel size and box size of either dataset 1 or 2. The output pixel size and the box size will be the same as the input reference map. After refinement, run CtfRefine with Estimate anisotropic magnification: Yes. This will refine the relative pixel size difference between two datasets. In the above example, the nominal difference is 10 %, but it might be actually 9.4 %, for example. Then run Refine3D again. The absolute pixel size of the output can drift a bit. It needs to be calibrated against atomic models.
How can I estimate the absolute pixel size?
Experimentally and ideally, one can use diffraction from calibration standards.
Computationally, one can compare the map and a refined atomic model. However, if the model has been refined against the same map, the model might have been biased towards the map. Also note that bond and angle RMSDs are not always reliable when restraints are too strong.