Lipid Monolayer Assay Protocol
by Brian J. Peter and Matthew K. Higgins
MRC Laboratory of Molecular Biology, Hills Road, Cambridge, U.K. CB2 2QH
If there are questions or comments, contact bpeter@mrc-lmb.cam.ac.uk or Harvey: hmm@mrc-lmb.cam.ac.uk
Background
Lipid monolayers have been used for many years as templates for the formation of two-dimensional crystals of soluble proteins (reviewed in (1)) and, more recently, membrane proteins (2). The principle of the assay is that phospholipids on an aqueous droplet adopt a conformation in which the hydrophobic tails point towards the air while the hydrophilic head groups contact the solution. Proteins of interest interact with the head groups and are concentrated in a two-dimensional array. A hydrophobic electron microscope grid interacts with the lipid tails, allowing the monolayer to be removed from the droplet and studied in the electron microscope. The lipid monolayer simulates the inner leaflet of the plasma membrane, and can be used to reconstitute early stages of endocytosis. The formation of ordered clathrin assemblies can be observed using negative stain electron microscopy, and platinum shadowing can reveal the invagination of these coats. For examples, see references (3,4). A schematic of the technique and a gallery of images obtained with this technique can be viewed using the links, or at http://www2.mrc-lmb.cam.ac.uk/groups/hmm/epsin/EM/
Reagents
HKM buffer (25mM Hepes pH 7.4, 125mM potassium acetate, 5mM magnesium acetate, 1mM dithiothreitol).
Chloroform
methanol
Phosphatidylinositol and Phospatidylinositol-4,5-bisphosphate (Avanti polar lipids) and were dissolved to 1mg/ml in 3:1 chloroform: methanol. Cholesterol (also Avanti) was dissolved to 10mg/ml in Chloroform. Phosphatidylserine, phosphatidylcholine and phosphatidylethanolamine (all from Sigma) were dissolved in chloroform to 10mg/ml. Lipid stocks were stored at -80°C.
2% uranyl acetate (Biorad) with 0.0025% polyacrylic acid (Sigma) in water (see note 2)
Purified clathrin. Clathrin should be dialyzed into HKM buffer and centrifuged for 20 minutes at 100000 gmax (e.g., 45000 r.p.m. in a Beckman TLA100 rotor) immediately prior to use to remove aggregates.
Purified AP180, epsin, or other clathrin- and lipid-binding protein.
Equipment
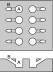
Teflon block with 60 microliter wells allowing for side injection (see figure 1)
Carbon and collodion-coated gold electron microscopy grids (e.g., G204G from Agar Scientific, coated first with collodion or formvar, and then with a thin layer of evaporated carbon)
Humid chamber, or covered container with a wet paper towel inside
Forceps for handling EM grids—self-locking spring forceps are especially useful
Whatman filter paper or similar, for blotting EM grids.
Parafilm
Vacuum evaporator, 0.2mm diameter piece of platinum wire (TAAB Laboratories),1mm thick tungsten wire (also TAAB) (necessary for platinum shadowing)
Transmission electron microscope.
Fig. 1. Top (above) and cross section (below) views of the teflon block used for monolayer formation. The block contains eight wells for processing samples in parallel. Main buffer well (A) should be 4 mm in diameter x 5 mm deep (or deeper) for a 60 microliter sample volume. The monolayer is formed on top of a buffer droplet in well A, and proteins and buffer are injected later through the side port B. See schematic diagram. 1.
Make up a lipid mixture containing 10% cholesterol, 40% PE, 40% PC and 10%
PtdIns(4,5)P2 to a final concentration of 0.1mg/ml in a 19:1 mixture
of chloroform: methanol (methanol is necessary to maintain PtdIns(4,5)P2
solubility). This mixture should be made on the day the monolayer is made. If
stored, it should be stored under argon at –80 degrees in a glass vial with a
glass or Teflon lid, for not longer than three days. 2.
Arrange teflon block in humid chamber, and fill wells of teflon block with HKM
buffer. Fill the wells with 40-60 microliters of buffer, such that the total
volume in the well will be 60 microliters after injection of protein samples.
See note 2. 3.
Carefully pipet (or inject with Hamilton syringe) 1 microliter of lipid mixture
on to the buffer in the well. As a negative control, inject pure chloroform
without any lipid (this will test for lipid dependence of any structures seen,
such as whether clathrin baskets
form in solution instead of clathrin coats on the monolayer surface).
See note 3. 4.
Incubate at room temperature for 60 minutes. The chloroform should evaporate,
leaving a monolayer of lipid on the surface of the buffer. 5.
Carefully place one EM grid, carbon side down, onto the top of each buffer
droplet. Grids should not be glow
discharged before use as a hydrophobic carbon film is required to adhere to the hydrophobic lipid tails
of the monolayer. 6.
Gently inject proteins into the side injection well. Final protein concentrations
in the well should be 0.5-2 micromolar for the AP180/epsin/adaptor protein, and
30-500 nanomolar for the clathrin. It is often useful to try several
concentrations, to account for differences in protein activity. 7.
Incubate 60 minutes at room temperature. 8.
Prepare Uranyl acetate stain. Lay a fresh piece of Parafilm on the bench, and
place two 30 microliter drops side by side on the Parafilm for each grid which will
be stained. Lay out a piece of Whatman filter paper to blot buffer and stain
from grids. Lay out a second piece of filter paper on which to set grids to air
dry. 9.
Gently inject approximately 30 microliters of buffer into the side injection
port; this will raise the grid up above the surface of the teflon block.
Immediately grab the grid with forceps and lift it vertically off of the
droplet. 10.
Blot the grid briefly by touching it to the filter paper, then touch it to the
first stain droplet and blot immediately. Touch the grid to the second stain
droplet, leave for 30 seconds, and blot briefly. This leaves a film of
stain on the surface of the grid in which the protein is embedded. If the grids will be platinum shadowed, hold
the grid to the filter paper for several seconds to ensure that the entire grid
surface dries. Lay the grid on
another piece of filter paper to dry. 11.
For negative stain EM, grids can be examined in the EM immediately. They are
also stable for several weeks, at least, at room temperature. 12.
If platinum shadowing is required, set up the vacuum evaporator with a
2cm long piece of platinum wire coiled tightly round a piece of 1mm thick
tungsten wire. Place the grids to be shadowed on a rotary platform at an angle
of 10° to the line between the platform
centre and the platinum coil. Create a vacuum in the evaporator. With a shield
between the grids and the wire, turn on the current to the tungsten wire. When
the platinum wire melts, remove the shield and the platinum will evaporate onto
the grids. For rotary platinum shadowing, start rotation of the platform was
immediately before removing the shield. For single angle shadowing, the
platform can remain stationary during the evaporation. 1-2 minutes of
evaporation is usually sufficient, but trials may need to be done to account
for differences in evaporators. This procedure will take approximately 3
hours. All reagents should be of the highest purity available, and buffers
should be filtered before use. 1. The addition of polyacrylic acid helps
to prevent the stain from precipitating and forming uranyl acetate crystals.
But this is not essential, alternatively UrAc solutions can be clarified by
filtration or centrifugation before use. 2. It is important to ensure that the
surface of the droplet is either flat or slightly concave. Overfilling the
teflon wells leads to a convex droplet surface, upon which the monolayer does not form properly. Also, filling
the wells with too little volume (less than 35 microliters, or depending on the
geometry of the block) can lead to an uneven surface near the side injection
port. 3. Trace lipid contamination (e.g.,
PtdIns(4,5)P2) on the teflon block can result in misleading negative controls.
Teflon block should be rinsed with hot water, then ethanol, and finally, soaked
overnight in a mixture of chloroform/methanol to remove any protein or lipid
residue. Hamilton syringes are also susceptible to trace lipid contamination. 1. Chiu W, Avila-Sakar AJ, Schmid MF.
Electron crystallography of macromolecular periodic arrays on phospholipid
monolayers. Adv Biophys. 1997;34:161-72. 2. Levy D, Mosser G, Lambert O, Moeck GS,
Bald D, Rigaud JL. Two-dimensional crystallization on lipid layer: A successful
approach for membrane proteins. 3. Ford MG, Mills IG, Peter BJ, Vallis Y,
Praefcke GJ, Evans PR, McMahon HT.
Curvature of clathrin-coated pits driven by epsin. Nature.
2002;419(6905):361-6. 4. Ford MG, Pearse BM, Higgins MK, Vallis
Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of
PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on
membranes. Science. 2001;291(5506):1051-5.
Procedure
Notes
References
J Struct Biol. 1999 Aug;127(1):44-52