Our structures
Our methods aim to open up new experimental possibilities in Structural Biology studies. By ourselves or through collaborations, we have used our methods to determine new structures. Some of these structures are described here. For an exhaustive list, please refer to the publications list.
TMEM106B filaments from brain
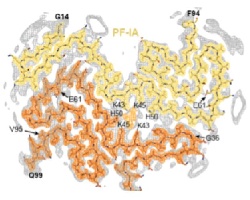
In close collaboration with Michel Goedert, we discovered amyloid filaments made of the transmembrane protein 106B (TMEM106B) in the brains of individuals with multiple neurodegenerative diseases, and solved their structures by cryo-EM. The presence of the same structures in older, but not younger, healthy control brains suggests TMEM106B filaments form in an age-dependent manner. Their relationship to disease is unclear.
Beta-amyloid 1-42 filaments from brain

In close collaboration with Michel Goedert, we determined atomic structures of amyloid-beta1-42 filaments from human brains. We identified two different protofilaments folds. Individuals with sporadic Alzheimer's disease have type I, whereas individuals with several other diseases have type II filaments in their brain. For the first time, we also identified an animal model that replicates filaments structures observed in human brain: the APP NL-F mouse brain contains type II amyloid-beta1-42 filaments.
Tau filaments from multiple tauopathies
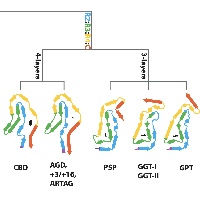
In close collaboration with Michel Goedert, we determined atomic structures of tau filaments from various 4R tauopathies, including progressive supranuclear palsy (PSP). Together with our previous tau structures and new ones from familial British and Danish dementia, these structures form the basis of a hierarchical classification of tauopathies that complements their clinical and post-mortem description, and allows identification of new diseases.
SARS-Cov-2 spike protein
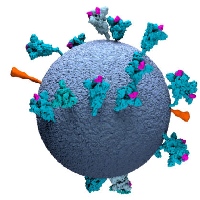
In response to the global COVID19 pandemic, and in close collaboration with the group of John Briggs, we contributed to structure determination of a thermostable variant of the SARS-COV2 spike protein for vaccine development, as well as the analysis of the SARS-Cov-2 spike on intact virions by electron tomography. Software developments in RELION-4 for subtomogram averages helped to obtain new insights in the tethering of the spike to the SARS-Cov-2 virion.
Apoferritin to atomic resolution

In collaboration with Abhay Kotecha at Thermo Fisher Scientific, we used a new cold field-emission gun, a new energy filter and a Falcon-4 camera to collect data on apoferritin for ~36 hours. Using the latest version of our RELION software, we were then able to calculate the first cryo-EM single-particle reconstruction to atomic resolution (1.22A). As expected, the map shows individual blobs of density for most atoms. Even hydrogen atoms are wellr esolved in difference maps with atomic models.
Synuclein filaments from MSA
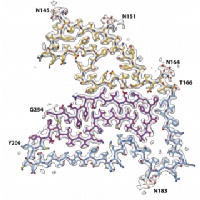
In close collaboration with Michel Goedert, we determined atomic structures of alpha-synuclein filaments from the brains of five individuals with multiple system atrophy. Two asymmetric filament types with four different protofilament structures were identified. These structures are different from previously determined filaments of recombinant alpha-synuclein. Based on 2D class averages of alpha-synuclein filaments from three cases of dementia with Lewy bodies, DLB filaments are also different. These results suggested that alpha-synuclein is similar to tau in these aspects.
Tau filaments from CBD
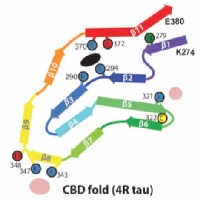
In close collaboration with Michel Goedert, we determined atomic structures of tau filaments from the brains of three individuals with corticobasal degeneration. The structures are different from those in Alzheimer's disease, Pick's disease and CTE. They incorporate a hydrophylic cofactor, which may be important for filament assembly in CBD.
,
Tau filaments from CTE

In close collaboration with Michel Goedert, we determined atomic structures of tau filaments from the brains of three ex-professional athletes with chronic traumatic encephalopathy (CTE): an American football player and two boxers. The structures are different from those in Alzheimer's disease. They incorporate a hydrophobic cofactor, which may be important for filament assembly in CTE. This also raised the possibility that cofactors may be important in determining the specificity of different structures in each disease.
Heparin-induced Tau filaments
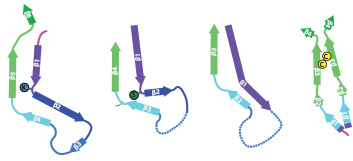
In close collaboration with Michel Goedert, we determined atomic structures of tau filaments that were assembled in-vitro using heparin. Assembly of tau filaments with heparin is widely used as an in-vitro model system for Alzheimer's filaments. However, our structures showed that heparin leads to more heterogeneous and smaller structures than those observed in disease.
Tau filaments from Pick's disease
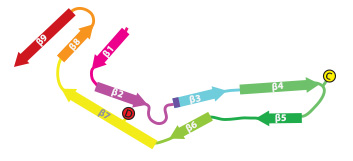
In close collaboration with Michel Goedert, we determined atomic structures of tau filaments from the brain of an individual with Pick's disease. This structure explains why in Pick's disease only tau isoforms with three microtubule binding repeat regions are incorporated into the filaments. The striking difference between the AD and Pick structures established the existence of molecular conformers of tau in different diseases.
Tau filaments from Alzheimer's disease
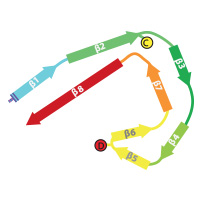
In close collaboration with Michel Goedert, we determined atomic structures of the paired helical and straight filaments of the tau protein that were purified from the brains of individuals with Alzheimer's disease. These structures reveal that residues 306-378 form the structured core of the filaments, and explain why in Alzheimer's disease a mixture of isoforms, containing both three and microtubule binding repeat regions, are incorporated into the filaments.
E.coli replisome
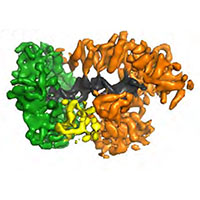
In close collaboration with Meindert Lamers, we determined a structure of the E.coli replisome comprising DNA polymerase III, clamp, exo-nuclease and tau, both with and without DNA bound. The structures reveal how a conformational change in the polymerase tail and tau may act as a molecular switch to change between processive DNA synthesis on the leading strand and discontinuous synthesis on the lagging strand.
Yeast U4/U6.U5 tri-snRNP spliceosome
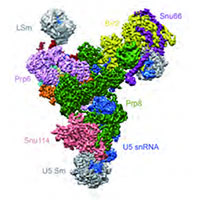
We assisted Kiyoshi Nagai in the determination of the structure of the tri-snRNP U4/U6.U5 spliceosomal complex from yeast to a resolution of 5.9 A. The resulting pseudo-atomic model reveals the essentially complete organization of its RNA and protein components, and provides crucial insights into the activation process and the active site of the spliceosome.
Human gamma-secretase
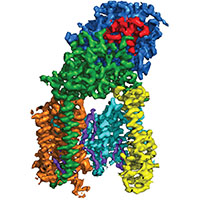
We determined the structure of human gamma-secretase in collaboration with Yigong Shi from Tsinghua University. With an ordered mass of only 130 kDa, this membrane complex was thought to lie beyond the scope of current cryo-EM techniques. However, careful imaging and (novel) statistical image processing by Xiaochen Bai resulted in a 3.4 Angstrom map, which revealed the structural organization of this complex.
Drosophila and human apoptosomes

We hosted Mengying Zhou, from Yigong Shi's lab at Tsinghua University for five months in our group to train her in cryo-EM structure determination. While she was with us, she solved several human apoptosome structures. Previously, Xiao-chen Bai in our lab had also solved a Drosophila structure as part of the same collaboration.
Rabbit ryanodine receptor
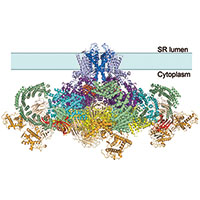
We determined the structure of the rabbit RyR1 ryanodine receptor to 3.8 Angstrom resolution, as part of a collaboration with Nieng Yan and Yigong Shi from Tsinghua University, Beijing. The near-complete atomic model explains the high ion conductance by ryanodine receptors and the long-range allosteric regulation of their channel activities.
P. falciparum ribosome bound to emetine and mefloquine
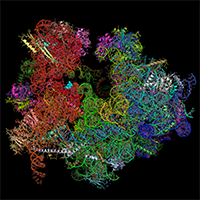
We hosted Wilson Wong, from Jake Baum's lab in Melbourne, Australia for five months in our group to determine the structure of the cytoplasmic ribosome of the malaria parasite: Plasmodium falciparum. By visualising this ribosome bound to emetine and to mefloquine, we learnt how these drugs work. These papers are an illustration of the potential of modern cryo-EM for structure-based drug design.
Mammalian ribosome:Sec61 complex
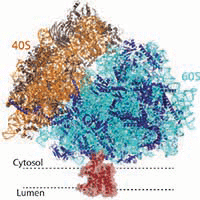
We assisted Manu Hegde in the determination of the structures of the mammalian ribosome-Sec61 complex in both idle and translating states to 3.4 and 3.9 Angstrom resolution, which permitted building of a near-complete atomic model of the mammalian ribosome, visualization of A/P and P/E hybrid-state tRNAs, and analysis of a nascent polypeptide in the exit tunnel.
Mitochondrial ribosomes
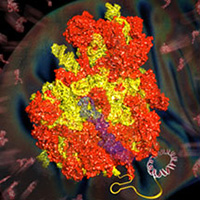
We assisted Venki Ramakrishnan in the determination of structures of both yeast and human mitochondrial ribosomes to near-atomic resolution. A true tour-de-force of de novo model building in the cryo-EM maps, aided by developments by Garib Murshudov and Paul Elmsley, revealed the unique architecture of ribosomal proteins and RNA in mitochondrial ribosomes compared to their cytosolic counterparts
Yeast ribosome:eIF5B complex
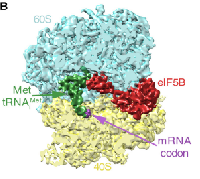
In collaboration with Venki Ramakrishnan, we determined the structure of a eukaryotic translation initiation complex. We used cryo-EM to efficiently guide our biochemical sample preparation, and finally isolated a subset of ~3% of the particles by image classification in RELION. The corresponding 5,000 particles still give a 6.5 Angstrom map.
3D DNA-origami object

In collaboration with Hendrik Dietz (Technical University Munich), we determined the structure of a 3D DNA-origami object that we designed to be particularly suitable for single-particle analysis. The structure is the first of its kind and shows that DNA-origami objects are more ordered than previously thought. In addition, the structure describes a range of non-natural DNA topologies to be mined for future use in nano-technology.