Structure and biophysics of major AMPA receptor complexes
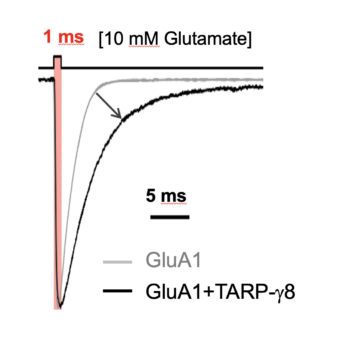
AMPAR activation by glutamate involves a spectrum of conformational transitions referred to as ‘gating’. These ultimately open the channel’s gate to permit cation flux and membrane depolarisation. Using a combination of electron cryomicroscopy (cryo-EM), patch-clamp electrophysiology, molecular dynamics simulations, and mass-spectrometry, we study:
- the organisation and stoichiometry of predominant AMPAR-auxiliary subunit complexes
- how auxiliary subunits influence transitions in the AMPAR gating cycle
- the identity, binding sites and role of AMPAR-associated annular lipids
- the development of therapeutic ligands targeting predominant AMPAR complexes (in collaboration with AstraZeneca and LifeArc).
Regulation of AMPA receptor organisation at potentiated synapses
A hallmark of learning (i.e. the encoding and storage of information) is the growth and potentiation of synapses. This process requires the recruitment of AMPARs into the post-synaptic density (PSD) and their accumulation into receptor nano-clusters. A central question is the nature of the machinery facilitating AMPAR organisation at potentiated synapses. To address this we:
- investigate essential AMPAR clustering components, through recordings of neurons expressing modified AMPAR complexes (expressed virally in genetically modified mice)
- image the sub-synaptic distribution of AMPAR varieties using 3D STORM and STED, prior to and after synapse potentiation (induced electrically or via 2-photon glutamate uncaging)
- identify AMPAR synaptic clustering/anchoring proteins using proximity labelling approaches
- re-constitute core clustering elements (identified through the above approaches) together with AMPARs in synthetic ‘hemi-synapses’