Replication associated epigenetic instability
Although all cells in our body contain the same genome, each cell type requires a different pattern of genes to be expressed. When cells divide the pattern of gene expression is maintained. The information that determines this pattern is not directly encoded in the DNA and is hence termed 'epigenetic'. The physical carrier of this memory is unclear and has been a subject of intense research, and a number of models have been put forward.
One of the most prevalent suggests a central role for the post translational modification of histone proteins by acetylation or methylation. These modifications have been associated with particular transcriptional states. For instance the presence of di- or tri-methylation of histone H3 (H3K9me2/3) is associated with regions of repressed transcription, while H3K9 acetylation (H3K9ac) and H3K4me3 is associated with transcriptionally active loci..
For histone marks to be epigenetic they must be propagated through replication. To understand this model it is first necessary to understand how histones are managed at the replication fork. For this purpose we can concentrate on the core of the nucleosome, which comprises a heterotetramer of two H3 and two H4 molecules. Nucleosomes are displaced from the DNA by the action of the replicative helicase. Current evidence suggests that the core H3/4 tetramers are then redeposited intact on one of the two newly synthesised daughter strands.
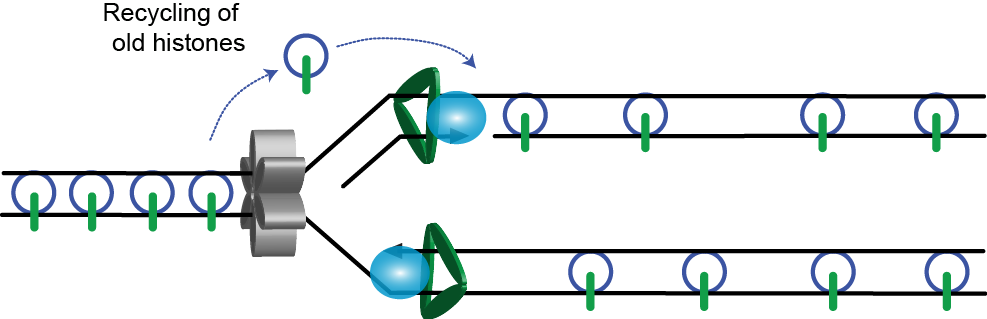
1. Parental histone recycling
Parental H3/4 tetramers (blue circles) and carrying post-translational modifications characteristic of that region of chromatin (green lines) are displaced ahead of the replicative helicase. They are distributed equally between the two daughter strands and are installed immediately behind the replication fork.
In order to maintain nucleosome density these parental histones must be combined with newly synthesised histones.
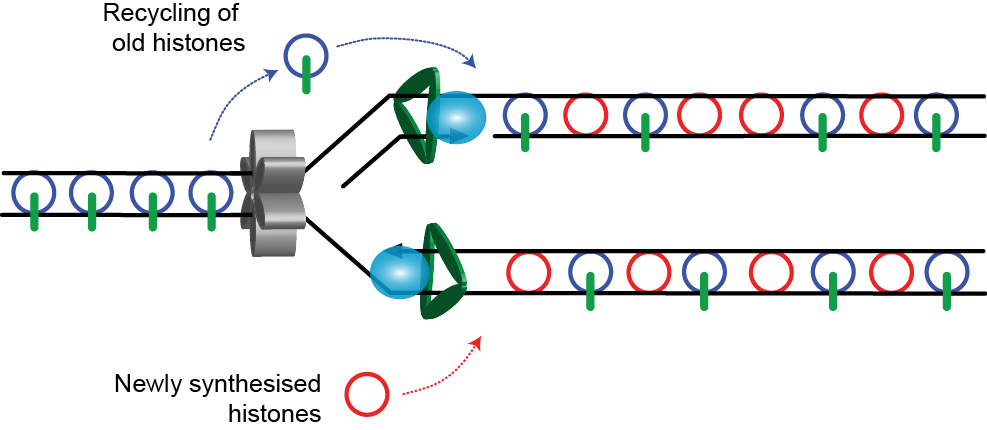
2. New histone incorporation
If only parental histones were available during replication, nucleosome density would be halved. Thus parental H3/4 tetramers are interspersed with newly synthesised H3/4. Most of the time, the old and new proteins are not mixed within the same nucleosome.
This generates a intermediate state in which parental, modified tetramers are mixed aproximately 50:50 with newly synthesised, unmarked tetramers. For the model to be complete the modifications on the parental tetramers need to be transferred to the naïve tetramers. For a number of modifications, particularly those associated with repressed chromatin such as H3K9me2 and H3K27me3, enzyme systems capable of doing this are known, for example HP1 can bind to H3K9me2/3 and recruit the histone methyltransferase SUV39 to methylate H3K9 on an adjacent nucleosome.
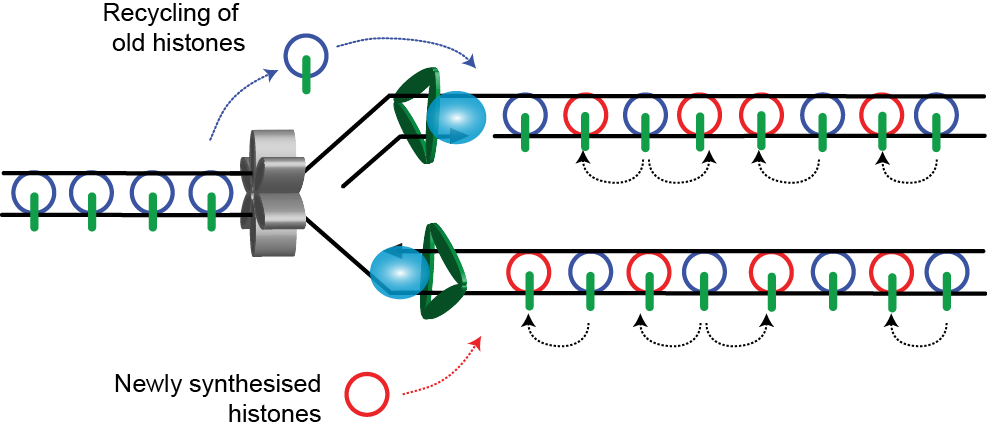
3. Transfer of marks to the new histones
The newly synthesised H3/4 lack parental marks characteristic of the chromatin being replicated, for example H3K9me3 or H3K27me3 if repressed or H3K4me3 if active resulting in a hypomodified state. Following replication, the parental marks are added to the newly synthesised histones, restoring the parental state. The mechanisms by which this happens are much better understood for heterochromatin than for actively expressed loci
If the modifications carried on parental H3/4 are truly epigenetic, i.e. they carry epigenetic information through the cell cycle, this model predicts that processes that uncouple DNA unwinding and histone displacement from DNA synthesis and re-chromatinisation should also disrupt the cell's ability to accurately maintain transcriptional states. This happens in cells lacking Rev1 where DNA damage results in increased used of post-replicative gap filling (see Mutagenesis and DNA damage tolerance)
We have shown that the Y family polymerase Rev1 is required for replicating naturally occurring structured DNAs called G quadruplexes. In DT40 cells lacking Rev1, certain normally repressed genes containing G quadruplex DNA sequences exhibit localised bias to new histone incorporation, loss of marks associated with transcriptional repression and increased transcription. Our data suggest that repeated replication arrest at G quadruplex structures in Rev1-deficient cells leads to uncoupling of DNA synthesis from histone recycling leading to failure to propagate an existing repressive chromatin environment.
We have gone on to show that several other factors can induce this form of replication-dependent epigenetic instability, including mutations in G4 helicases (e.g. FANCJ), G4 stabilising ligands, a failure of repriming due to loss of PrimPol and, importantly, induction of global replication stress.
4. Replication impediments cause epigenetic instability
The newly synthesised H3/4 lack parental marks characteristic of the chromatin being replicated, for example H3K9me3 or H3K27me3 if repressed or H3K4me3 if active resulting in a hypomodified state. Following replication, the parental marks are added to the newly synthesised histones, restoring the parental state. The mechanisms by which this happens are much better understood for heterochromatin than for actively expressed loci
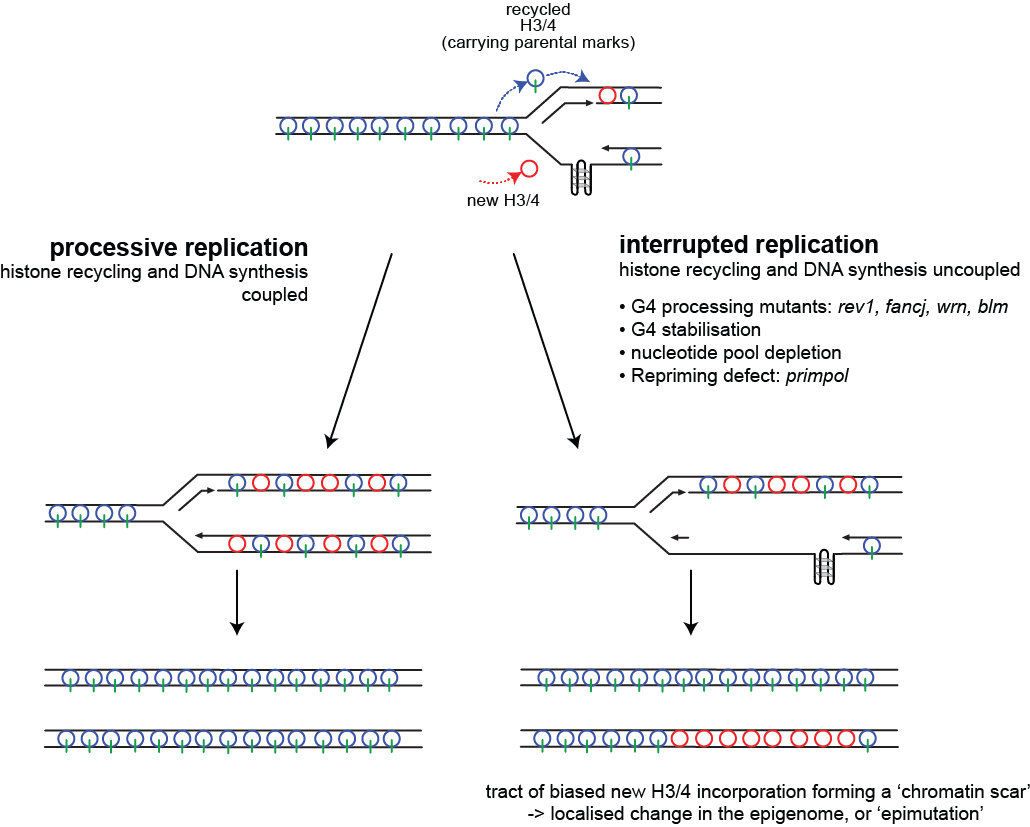
These data thus provide support for the importance of histone recycling for maintenance of at least some epigenetic states and suggest a novel mechanism for how a genetic change could give rise to widespread epigenetic changes, a feature of many cancer cells.