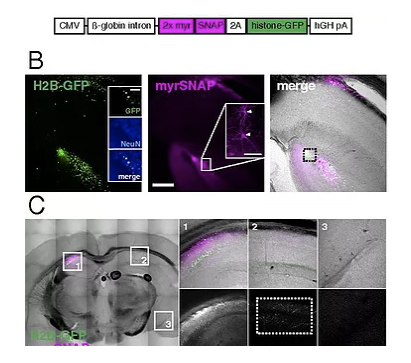
Tools for Connectomics and Neurogenetics
We are always actively engaged in developing new viral and non-viral tools to map neural circuits connectivity and manipulate circuit function.
SiR
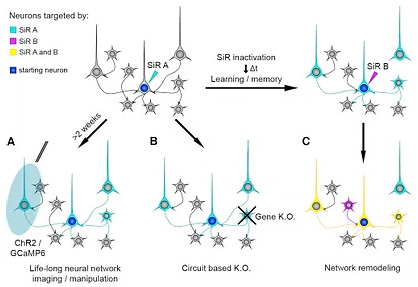
Neural networks, circuits of neurons, are emerging as the fundamental computational unit of the brain and it is becoming progressively clearer that neural network dysfunction is at the core of a number of psychiatric and neurodegenerative disorders. Yet our ability to target and study specific neural networks remains limited. Until now Rabies virus, which can jump synapses, has been used to investigate neural networks. However, the virus causes neuronal death resulting in a relatively short time window in which the networks can be studied. Our work resulted in a novel approach which provides, for the first time, a life-long ability to study neural networks, without affecting their normal physiology.
We designed a self-inactivating Rabies virus (SiR), which retains the ability to jump synapses, thus reaching the entire neural network, but does not cause neuronal death. During the initial viral infection, the neurons in the network are permanently labelled and genetic manipulation is made possible. The virus can modify the neurons so they constantly produce a fluorescent protein, allowing the network to be visualised. Crucially, we modified the virus by adding a tuneable switch – the SiR contains selected proteins which can be targeted for degradation. Without these proteins, the virus is unable to replicate and switches off after the initial infection, thus avoiding death of the neurons and leaving them permanently labelled but virus-free.
Progress in science often depends on new techniques and with this novel approach it is now finally possible to genetically manipulate neural networks and follow their behaviour over prolonged periods of time. This will be important at a basic science level to understand how the brain works, and excitingly will also offer insight into how neural networks change upon experience and learning. In addition, this new technique will pave the way to the engineering of the genetic content of selected neurons within a network, which would be extremely useful in neurodegenerative and psychiatric disorders where disease genes could be targeted, with therapeutic potential.
Paper in Cell.
You can aslo look at some of the commentaries on this work :
- Spotlight Article: Menegas, M., Uchida, N. and Watabe-Uchida, M. Trends in Neuroscience, 2017 Sept 7; 40 (10): 589–591
- Research Highlight: Irene Jarchum. Nature Methods, 14, 836, 2017
- Selected F1000
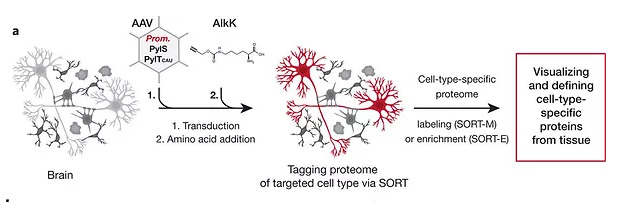
Labeling and identifying cell-specific proteomes in the mouse brain
A small contribution from our lab to the work of the group of Jason Chin to implement a viral expression system for the cell-type-specific tagging of proteomes in the brain. The strategy relies on the stochastic orthogonal recoding of translation (SORT) method previously developed by Jason’s lab. The approach is based on viral-mediated expression of an orthogonal pyrrolysyl-tRNA synthetase-tRNAXXX in the desired neuronal type and allows to label or enrich the proteome of genetically defined neuronal classes.
Paper in Nature Biotechnology.
Ultrafast tissue staining with chemical tags
Our small contribution to the work of the group of Greg Jefferis for the development of viral vectors for the delivery of genetically encoded chemical tags that result in rapid, even staining of thick biological samples with high-signal and low-background labeling. This tag-based approach drastically improves the speed and specificity of labeling genetically marked cells in intact and/or thick biological samples.
Paper in PNAS