EHD Home Page |
What are EHDs?
Eps15 homology domain-containing proteins are a family of membrane remodelling proteins which are related to the Dynamin Superfamily of large GTPases.
-Highly conserved in all higher eukaryotes, but not in
yeast and bacteria
-Four paralogues in human, 70 - 80% amino acid identity, all with different localisations
The pictures and movies below have been made to accompany our paper on EHD2 (Nature 3Oct2007) and thus this web page aims to give a brief introduction and to display useful movies that illustrate points in the paper.
(Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling, Daumke et al doi: 10.1038/nature06173) |
Where do EHDs work in the cell?
They function in MHC class I recycling (Caplan EMBO J 2002), in exit from the endocytic recycling compartment (Grant NCB 2001), and in endocytosis of activated TrkA receptors (Valdez et al J. Neurosci 2005).
Many of the functions likely remain undiscovered.
|
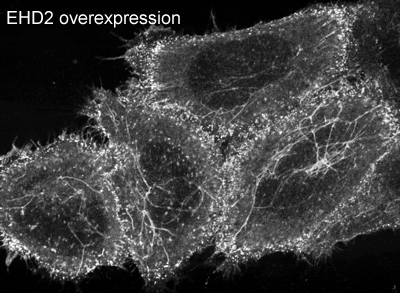
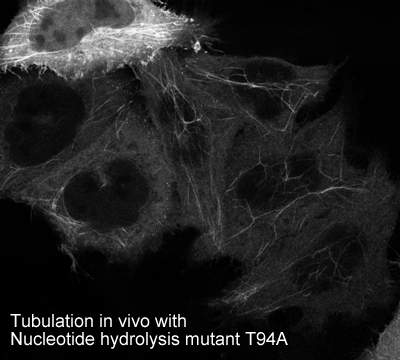
EHD2 is localised largely to puncta close to the plasma membrane. In overexpressing cells we also observe EHD2 positive tubules |
EHDs are Dynamin Superfamily Members: Like other members of the superfamily EHDs:
-have a low affinity for nucleotides (low micromolar range)
-bind to negatively charged membranes (EHD2 shows Ptd Ins(4,5)P2 preference)
-tubulate membranes in vivo when hydrolysis deficient mutants are overexpressed
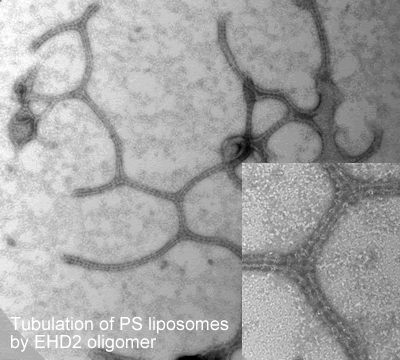
-tubulate liposomes in vitro forming ring-like structures
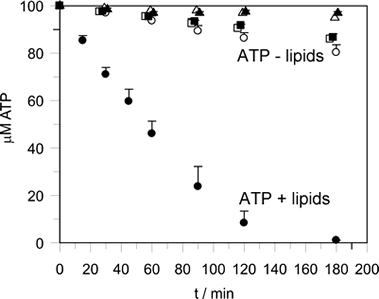
-show lipid-stimulated nucleotide hydrolysis on membrane binding |
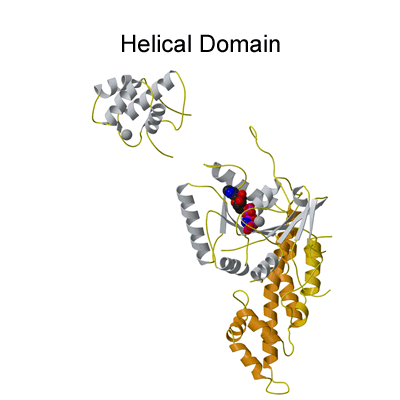
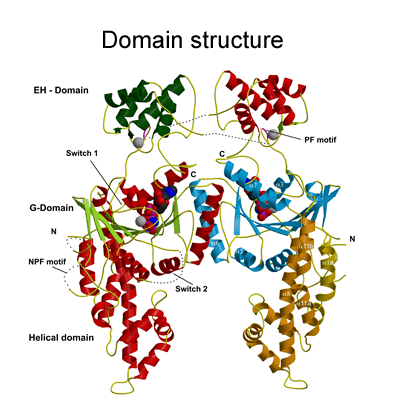
Structure of the EHD2 dimer
The EHD2 structure solved was a dimer (see Figure 2a) and the movies below are to complement the description given in the paper so that the reader can gain a better 3-dimensional impression. |
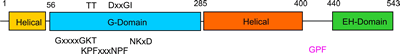 |
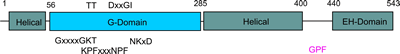
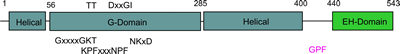
The G-domain binds and hydrolyses ATP, while the helical extension forms the platform for membrane binding and the EH domain is a protein interaction module (binding to NPF-like peptides) |
|
|
The dimerisation interface seen in the EHD2 structure has not been seen before for dynamin superfamily members (see supplementary information in paper), but the conserved surface (coloured purple above) is the same face used for oligomerisation in Guanylate Binding Protein (GBP), bacterial dynamin and septins, and is likely the interface used for EHD oligomer formation on membrane binding (see link for comparison of oligomerisation interface with those of GBP, BDLP and septins). |
 |
 |
 |

 |
How do we think EHDs work? EHD2 dimers interacts with membranes via ionic interactions mediated by a highly curved interface and we predict that this interaction results in the insertions of V321 and F322 into the hydrophobic phase of the lipid bilayer. Both the curved interface and the hydrophobic residue insertions will result in the bending of the membrane towards the EHD2 dimer. The membrane curvature imposed by our proposed oligomer would be perpendicular to the curvature imposed by the concave membrane binding face of the EHD2 dimer. This would cause considerable curvature stress in the lipid bilayer and would thus facilitate the lipid rearrangement required for the formation of intermediate stages towards membrane fission/fusion. Finally we observe that nucleotide hydrolysis is most likely leading to membrane scission in vivo, and thus we would argue that conformational changes induced by nucleotide hydrolysis are leading to further membrane destabilisation. |
|
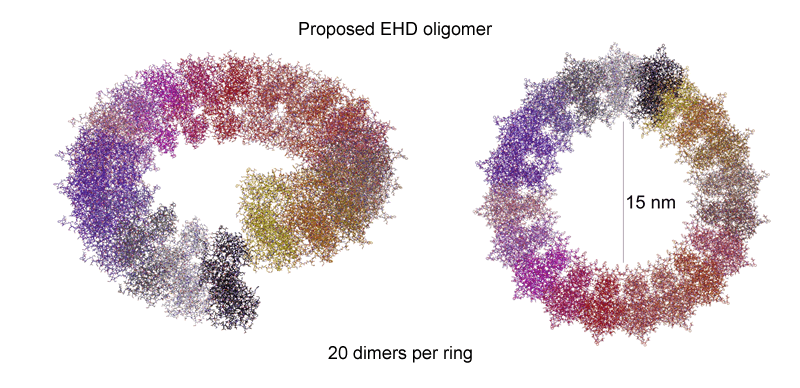 |
Oligomerisation in vitro
 |
Oligomerisation in vivo
 |
|
Proposed movement of EH domain belonging to light blue EHD monomer on oligomerisation (see figure 4 in paper). On interaction of the orange dimer and the blue dimer (light and dark blue) the EH domains get displaced from the superjacent positions. Only the light blue EH domain is illustrated, leaving its position on top of the dark blue monomer and binding to the side site PF motifs on an orange monomer, helping to organise the oligomer and making the EH domain inaccessible to other potential ligands.
The fit of the oligomer can be seen in the movies below, where the EH domains have been removed for clarity (see also figure 4 in paper) |
|
|
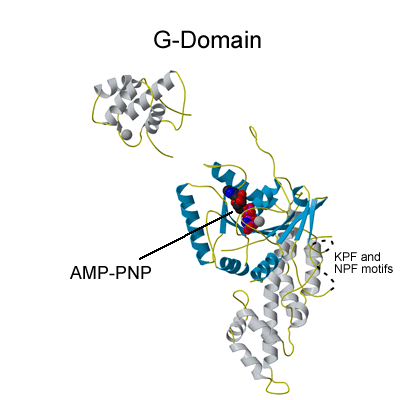
Nucleotide Specificity?
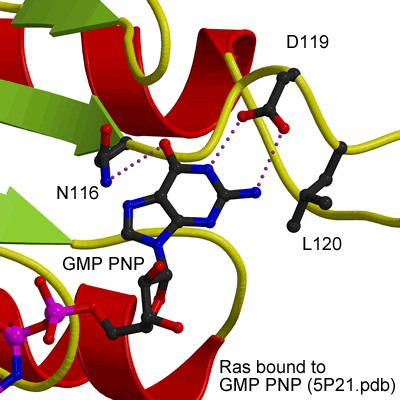
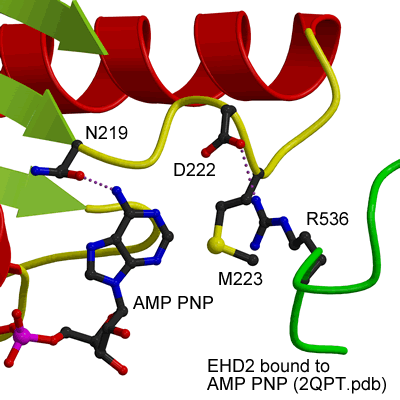
EHDs bind and hydrolyse ATP, despite having a G-domain that normally binds to GTP (Figure 1). This G-domain is highly conserved with Ras and Dynamins and the structural explanation of ATP binding is shown in the figure below (see supplementary figure 4 for more details).
ATP hydrolysis is stimulated by membrane binding. In the dimer structure the Switch I and II regions are not well ordered, and it is assumed that on oligomerisation (upon membrane binding) that these regions become ordered and hydrolysis is thus stimulated by positioning of a catalytic residue.
|
Lipid-stimulated ATP hydrolysis
|
GTP binding site of Ras
 |
ATP binding site of EHD2
 |
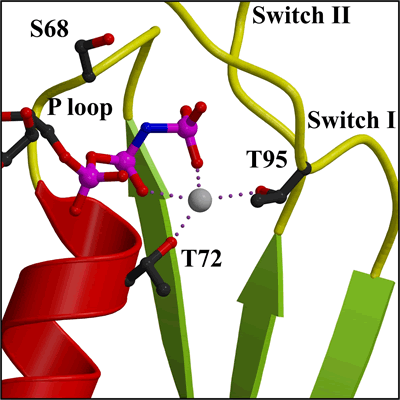 |
Detail of Mg2+ in nucleotide binding site (closer view of area in Figure 2a) |
Apposition of nucleotides in oligomer
In the trimer of dimers shown here, one can see that nucleotides from diagonally opposed monomers are in closest proximity. This arrangement may well be important in the catalytic mechanism (see also Figure 4c). |
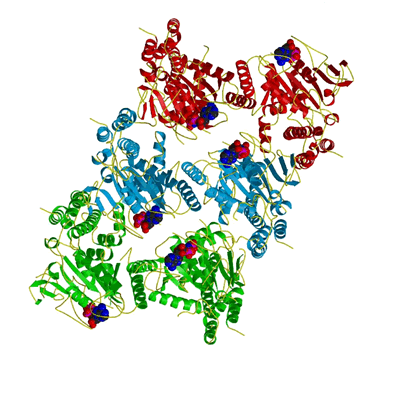 |
Lipid Specificity?
EHDs bind weakly to most negatively-charged lipids but bind very tightly to PIP2 (PtdIns(4,5)P2). |
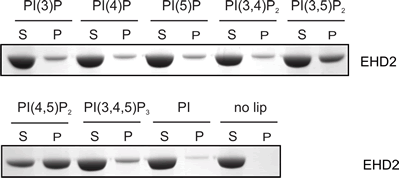 |
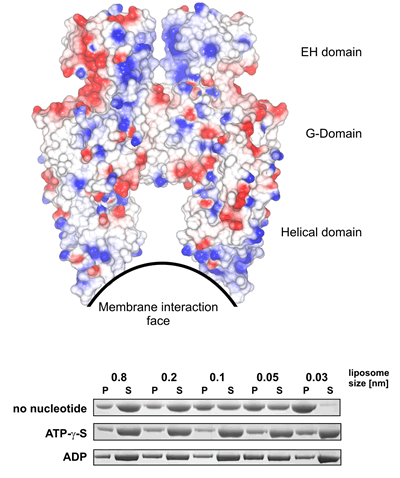 |
Curvature Sensitivity? (see Figure 3a) In the absence of nucleotides (when the protein does not have as high a propensity to form oligomers) EHDs bind preferentially to very highly positively curved membranes (just like the shape of the membrane interaction site of the dimer). However in the presence of nucleotides there is very little curvature sensitivity (likely because the oligomer is able to drive its own curvature preference). |
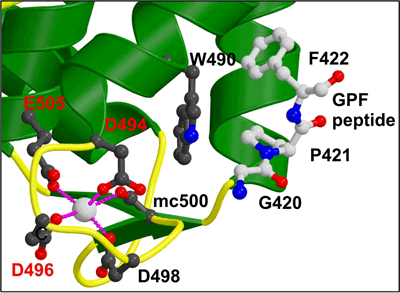 |
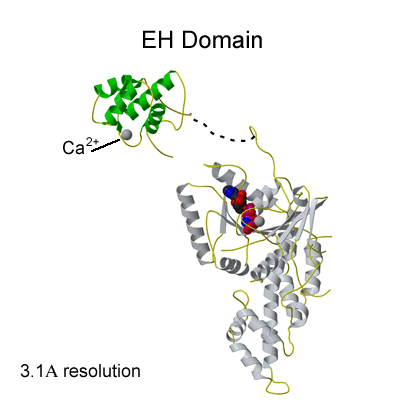
Binding of NPF peptides to EH domain
A hydrophobic pocket accommodates the F of the NPF peptide, formed by W490 of the EHD2 EH domain. The calcium ion of the EH domain is coordinated by 4 acidic residues and by a main chain carbonyl group. This is the first crystal structure of an EH domain bound to a peptide. Structural comparisons are shown in Figure 2c. |
Major questions that should still be answered:
A. How does oligomerisation effect nucleotide hydrolysis?
B. Is there a conformational change in the dimer or the oligomer on nucleotide hydrolysis?
C. Does one need a ring or a helix for activation of nucleotide hydrolysis and function?
D. What do these proteins do in vivo?
Our publications on EHDs
Daumke et al Nature, 2007. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Published online on 3Oct2007 (doi: 10.1038/nature06173 (see papers) (see nature link). |