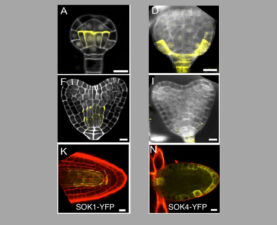
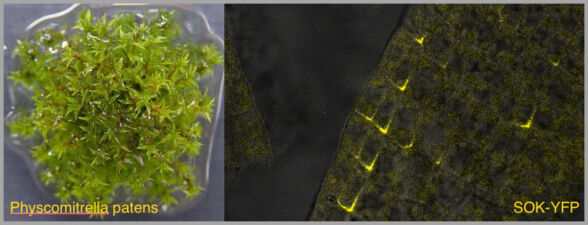
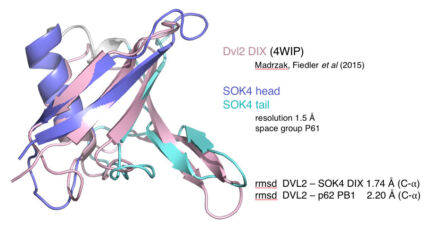
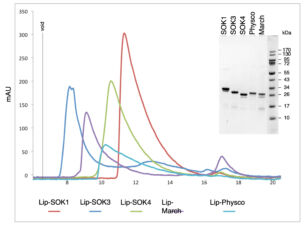
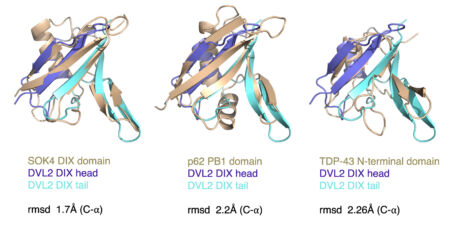
We have recently started two different plant collaborations to explore whether the mechanism of dynamic head-to-tail polymerisation might be conserved beyond animals. One collaboration was with Dolf Weijers (Wageningen, Netherlands) who discovered the first plant protein showing a polar localisation within Arabidopsis cells, called SOSEKI. SOSEKI proteins play a role in cell polarity and are highly conserved in all land plants including mosses and lichens. They contain a bona fide polymerising DIX domain in their N-termini (called DOX), similarly to animal DIX and DAX. Despite any obvious primary sequence conservation between the two domains, DOX turned out to be the closest structural relative of DIX, as judged by the highly similar backbones of their folds in their crystal structures. Remarkably, the DOX domain can be traced back to unicellular organisms, where its function may be in processes other than planar cell polarity that require phase separation.
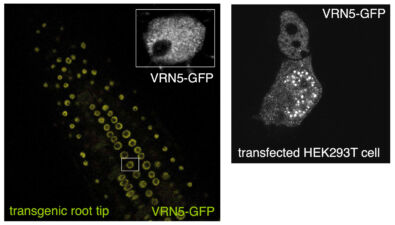
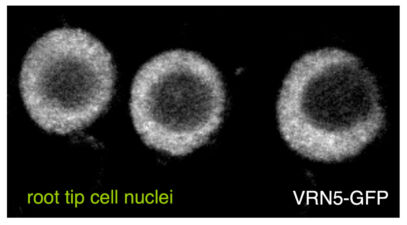
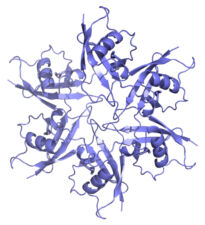
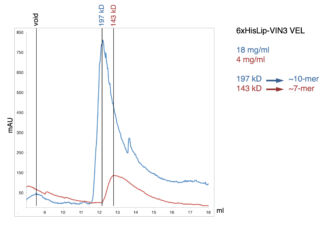
Another collaboration was with Caroline Dean (John Innes Centre, Norwich) on the VEL proteins, which control flowering of Arabidopsis plants after a cold spell (called vernalisation), by assembling heritable Polycomb-dependent silencing complexes at multiple target genes. These proteins associate with chromatin through an unusual PHD domain without histone-binding activity. They also contain a polymerising VEL domain whose structural fold resembles an atypical four-helix bundle. Its polymerising activity is essential for vernalisation in the case of VIN3, the cold-induced VEL paralogue which plays a key role in initiating Polycomb silencing, but not in the case of VRN5 which is functionally and molecularly distinct from VIN3. The VEL domain represents only the third known structural fold engaging in dynamic head-to-tail polymerisation – in addition to the SAM and DIX domain families. These plant projects have highlighted a deep conservation of what turned out to be a fundamental and widespread mechanistic principle – namely that multi-protein complexes can be assembled through dynamic head-to-tail polymerisation, which transiently increases the binding avidity of their components to effector proteins
Relevant references:
- Franco-Echevarría, E., Rutherford, T.J., Fiedler, M., Dean, C., Bienz, M. (2022)
Plant vernalization proteins contain unusual PHD superdomains without histone H3 binding activity.
J Biol Chem 298(11): 102540 - Fiedler, M., Franco-Echevarría, E., Schulten, A., Nielsen, M., Rutherford, T.J., Yeates, A., Ahsan, B., Dean, C., Bienz, M. (2022)
Head-to-tail polymerization by VEL proteins underpins cold-induced Polycomb silencing in flowering control.
Cell Rep 41(6): 111607 - Franco-Echevarría, E., Nielsen, M., Schulten, A., Cheema, J., Morgan, T.E., Bienz, M., Dean, C. (2023)
Distinct accessory roles of VEL proteins in Polycomb silencing.
Genes Dev 37(17-18): 801-817
- Schulten A, Jang G-J, Payne-Dwyer A, Nielsen ML, Bienz M, Leake MC & Dean C (2024)
Functional specialization of Arabidopsis VEL polymerization domains in the switch to Polycomb silencing.
bioRxiv