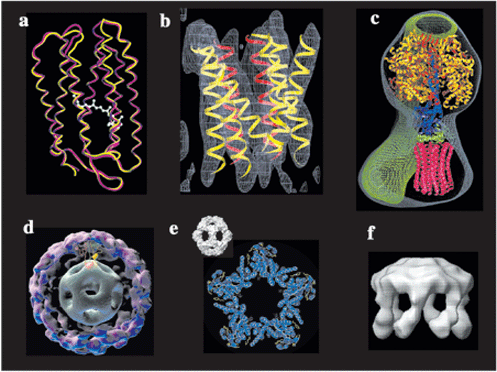
a. Two principal conformations of bacteriorhodopsin - Subramaniam, S. & Henderson, R. Nature 406, 653-657 (2000). Molecular mechanism for vectorial proton translocation by bacteriorhodopsin.
b. EmrE - Ubarretxena-Belandia I, Baldwin JM, Schuldiner S & Tate CG EMBO J. 22: 6175-6181 (2003). Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer
c. F1Fo-ATP synthase - Rubinstein, J.L., Walker, J.E. & Henderson, R. EMBO J. 22, 6182-6192 (2003). Structure of the mitochondrial ATP synthase by electron cryomicroscopy.
d. Pyruvate Dehydrogenase E1.E2 - Milne, J.S., Shi, D., Rosenthal, P.B., Sunshine, J.S., Domingo, G.J., Wu, X., Brooks, B.R., Perham, R.N., Henderson, R. & Subramaniam, S. EMBO J. 21, 5587-5598 (2002). Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine.
e. E2CD, surface shaded and surface net plus backbone - Rosenthal, P.B. & Henderson, R. J. Mol. Biol. 333, 721-742 (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single particle electron cryomicroscopy.
f. Neurospora H+-ATPase - Rhee, K.-H., Scarborough, G.A. & Henderson, R. EMBO J. 21, 3582-3589 (2002). Domain movements of plasma membrane H+-ATPase: 3D structures of two states by electron cryo-miocroscopy.
