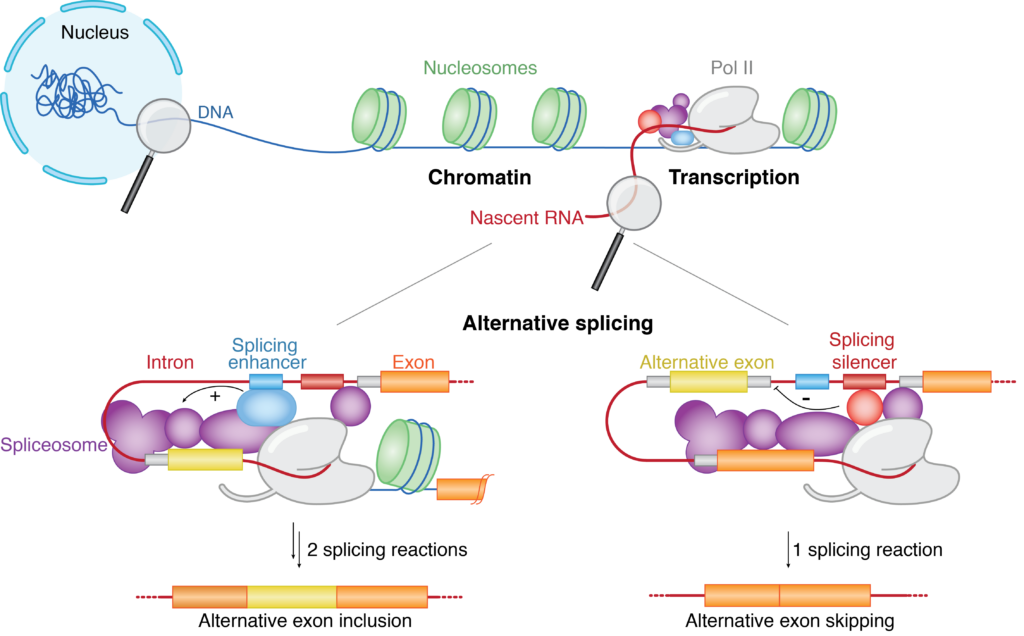
Splicing of precursor messenger RNA (pre-mRNA) is a ubiquitous hallmark of gene expression in all eukaryotes. Within the cell, splicing occurs simultaneously with pre-mRNA synthesis by RNA polymerase II (Pol II) and is mechanistically coupled to transcription. The choice of the splice sites is not always unique, as a single gene can give rise to multiple functionally distinct mRNA isoforms through alternative splicing. In fact, more than 95% of human genes are alternatively spliced, massively expanding the coding potential of our genome. Dysregulation of alternative splicing contributes to cancer pathogenesis and is under study as a biomarker of diseases. Our research aims to understand the mechanism governing alternative splice site selection and the crosstalk between the transcription and splicing machineries.
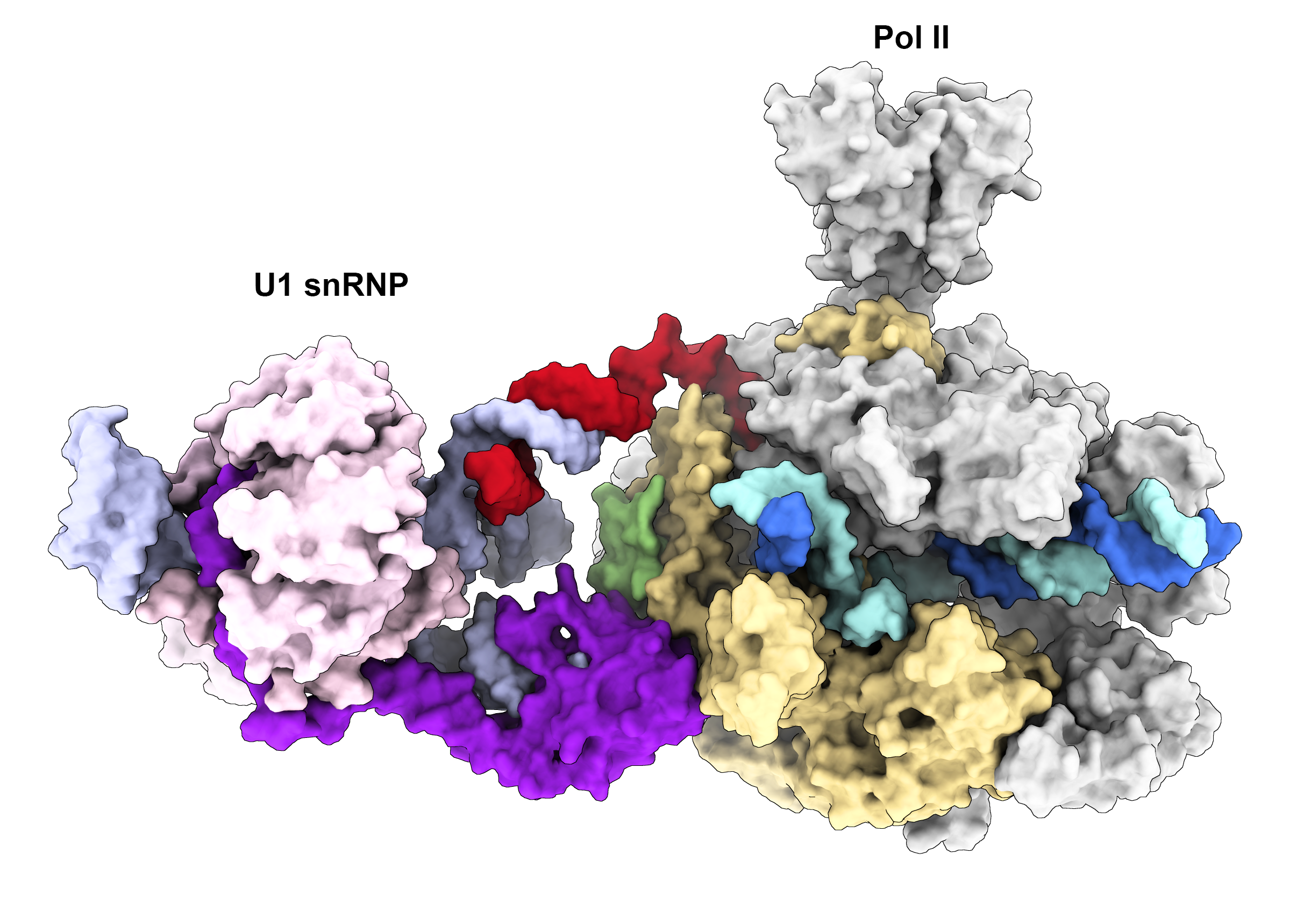
We recently determined the cryo-EM structure of the transcribing Pol II in complex with U1 snRNP, the first spliceosomal building block that engages pre-mRNA. The structure revealed for the first time a direct protein interaction between the transcription and splicing machineries within a large supercomplex and provided insights into how functional pairing of distant intron ends and spliceosome assembly can occur on the Pol II surface.
Our lab employs a multidisciplinary approach combining biochemical reconstitution, functional assays, structural biology and in vivo RNA sequencing to define the molecular basis of transcription-coupled alternative splicing. Our long-term research goal is to understand how the transcription and splicing machineries functionally interact to regulate gene expression.