Comparisons of close structural relatives |
||
First assemble a really good sequence alignment with the sequence of a known structure. This can be done using web servers with either global or local alignment preferences. Because the BAR proteins are largely alpha-helical it is helpful to know where the helices begin and end and thus a server that takes this into account will give a more accurate alignment. We use HHpred. The results of HHpred can be uploaded directly into Modeller. In choosing which structure to use, be careful, because some of the solved structures are slightly distorted due to crystal-crystal contacts. It is likely that one is attempting a comparison with a known structure in order to be able to know which residues should be mutated to abolish ligand/membrane/dimerization interface interactions. These residues can already be assumed from a good alignment and thus threading may be an overzealous exercise at this point. |
||
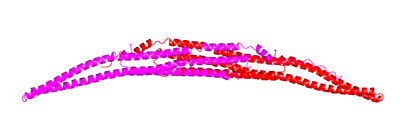
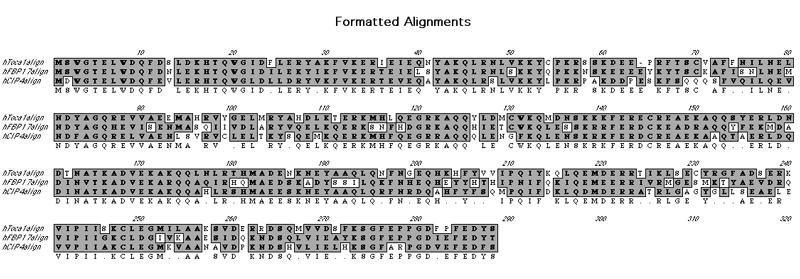
Toca1, FBP17 and CIP4 subfamily |
||
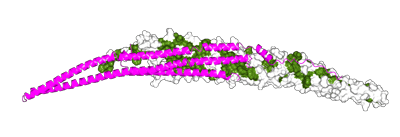
Left monomer is FBP17 and right monomer is Toca1. Toca1 sequence has been threaded using Threader and displayed using CCP4mg. The hydrophobic nature of the dimer interface is displayed in grass-green.
|
 |
|
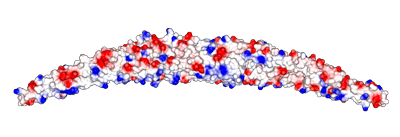
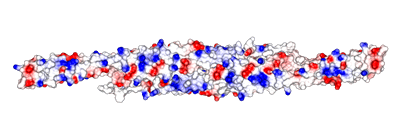
Surface electrostatic potential are also shown for the orientation above and also for the concave surface. It is interesting that there is not a strong concentration of positive charged residues at the extremities of the F-BAR like in some BAR proteins. |
 |
|
Click on images for higher resolution versions. Toca1 pdb |
 |
|
 |
||
 |
||
Back to BAR-Superfamily home