|
Structure of a BAR module
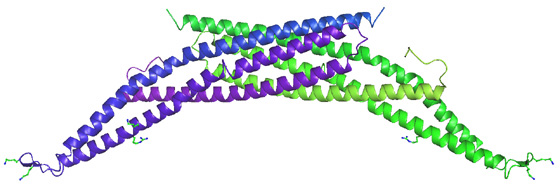
The BAR (Bin/Amphiphysin/Rvs; pfam03114 and SMART00721) domain is the most highly conserved feature of amphiphysins from yeast to humans. In the amphiphysin BAR structure below, two BAR domains form a crescent shaped homodimer with positively charge residues distributed along the membrane-interacting concave face. We refer to the homodimer as a BAR module
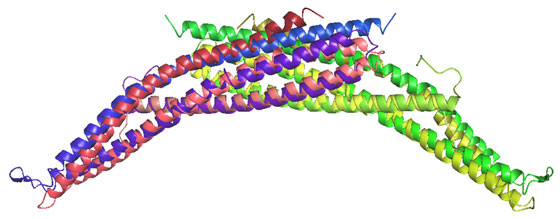
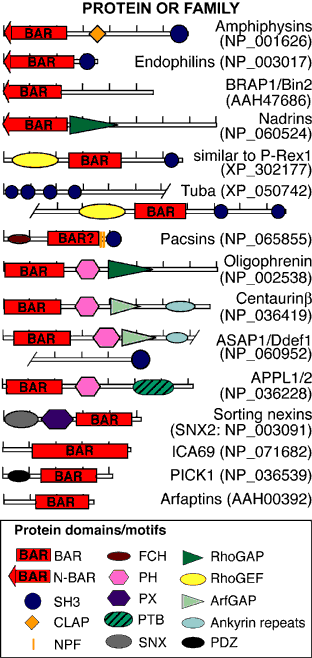
Unexpectedly, there was structural similarity to arfaptin2 (sequence identity 15%). We found that arfaptin2 also binds and curves membranes and is another member of the BAR family. What is an N-BAR domain N-BAR domains are BAR domains with an N-terminal amphipathic helix. Amphiphysin has such a helix, and initially in the absence of membranes it is unstructured but in the presence of membranes it folds and inserts into the bilayer. The consequence of insertion is that there is further induction of curvature as this works like a 'wedge' in one leaflet of the bilayer. This curvature can now be stabilised by the BAR module. For further discussion see McMahon and Gallop 2006 (Nature review) and Gallop et al 2006 (EMBO J). See artist's impression of membrane bending by BAR domains. BAR domains in other proteins Using the structural alignment, we searched the sequence database and found BAR domains in many proteins (see results of Hidden Markov Model searches). Some are described in the diagram below, ordered by their homology to amphiphysin (top) or arfaptin (bottom). Many of these proteins (N-BARs) have an amphipathic helix directly N-terminal to the BAR, and these proteins drive membrane curvature in vivo and in vitro. Other BAR domains are found in conjunction with PX or PH lipid binding domains, and in this context the BAR is sensitive to membrane curvature, preferring highly curved membranes. This curvature sensing effect is also found with the BARs alone, and is probably due to the curved shape of the dimer. We tested BAR domain functions in 3 amphiphysins, arfaptin2, nadrin2, centaurinBeta2, and oligophrenin1. We conclude that BAR domains are dimerisation, membrane binding, and curvature sensing modules found in many protein families.
Domain descriptions: BAR, Bin/Amphiphysin/Rvs; N-BAR, BAR with an N-terminal amphipathic helix; SH3, Src Homology 3; CLAP, Clathrin/Adaptor protein binding; NPF, sequence motif which binds Eps15; FCH, Fes/CIP4 homology; PH/PX, pleckstrin homology/phox homology phosphoinositide binding domain; SNX, N-terminal domain in sorting nexins 1 and 2; RhoGAP, GTPase activating regions towards Rho/Rac/Cdc42-like GTPases; RhoGEF, Guanine nucleotide exchange factor for Rho/Rac/Cdc42-like GTPases; ArfGAP, GTPase activating region for Arf GTPases; Ankyrin repeats, widespread protein-protein interaction motifs; PDZ, protein-protein interaction domain. In the diagram hatchmarks indicate 100 AA intervals, and accession numbers for human examples are in parentheses. For definitions of conserved domains and alignments see the NCBI conserved domain database Peter, B.J., Kent, H.M., Mills, I.G., Vallis, Y., Butler, P.J.G., Evans, P.R. and McMahon, H.T. (2003) BAR Domains as Sensors of Membrane Curvature: the Amphiphysin BAR Structure. published online on Science Express Nov 26; print version Jan 23, 2004.(abstract) Supporting online material.(pdf) Back to BAR-Superfamily Home Page |