e have developed methods that allow us to use the properties of genetically encoded unnatural amino acids to directly, specifically and synthetically observe and control molecular functions of user-defined proteins, with high spatial and temporal precision, in living cells and organisms. The overarching goal of these approaches is to bring a level of molecular, spatial and temporal precision – more typically associated with in vitro biochemistry – to the study of complex biological systems in their native, in vivo, context.
Central to the development of these approaches, which are described in more detail in the sections on photochemical genetics and protein labeling and imaging, is the ability to site-specifically direct unnatural amino acids into defined sites of proteins in the host system of interest using orthogonal synthetase/tRNA systems that recognize useful unnatural amino acids and direct their incorporation in response to the amber stop codon. We have developed approaches to direct the incorporation of unnnatural amino acids into protein in E. coli, S. cerevisiae, mammalian cells, and, most recently, animal models including C.elegans and D. melanogaster.
Genetic code expansion: from E. coli to yeast and mammalian cells. The pyrrolysyl-tRNA synthetase tRNACUA pair is orthogonal with respect to the endogenous synthetases and tRNAs in E. coli and we have shown that it can be evolved in the laboratory to direct the incorporation of a range of unnatural amino acids that are useful for in vivo studies. The pyrrolysyl-tRNA synthetase/tRNACUA pair is orthogonal not just to the synthetases and tRNA in E. coli, but also to the synthetases and tRNAs in yeast and mammalian cells. PylRS can, therefore, be evolved for new amino-acid specificity in E. coli and then used in eukaryotic cells. While the PyltRNACUA sequences used in E. coli can also be used in yeast and mammalian cells, the regulatory elements used to direct the transcription of the heterologous tRNA required extensive optimization.
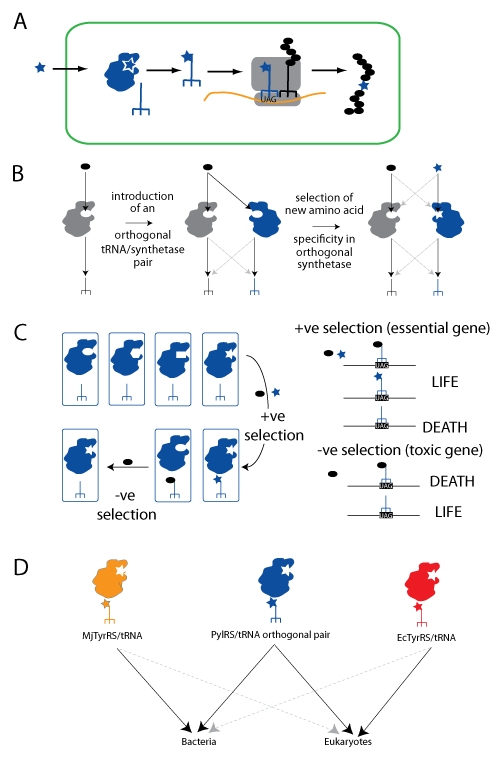
Expanding the Genetic Code using the PylRS/tRNACUA pair. A. An unnatural amino acid (blue star) is taken up by the cell. It is specifically recognized by an orthogonal aminoacyl-tRNA synthetase and attached to the orthogonal amber suppressor tRNACUA (blue trident), which is decoded on the ribosome in response to an amber codon. Natural amino acids are shown as black ovals. B. Orthogonal synthetase tRNACUA pairs are generated in two steps: import of a heterologous tRNACUA into a host containing a set of natural synthetases (grey) that use natural amino acids, and the subsequent selection of a mutated active site in the orthogonal synthetase to recognize the unnatural amino acid. C. A large library of active site variants of the synthetase is subject to positive selection for activity with either natural or unnatural amino acids, by virtue of their ability to suppress a stop codon in a gene essential for survival. Synthetases using natural amino acids are subsequently removed by a negative selection in the absence of the unnatural amino acid in which they lead to suppression of a stop codon in a toxic gene leading to cell death. D. The PylRS/tRNACUA pair is orthogonal and can be used in both bacteria and eukaryotic cells. The MjTyrRS/tRNACUA and the EcTyrRS/tRNACUA pairs have both been used for genetic code expansion, but they are only orthogonal in prokaryotic or eukaryotic cells respectively.
Eukaryotic tRNA genes contain RNA polymerase III promoter sequences internal to the structural tRNA gene. The sequence of the promoter is, therefore, intimately associated with the structure and function of the tRNA. As the tRNAs we import into eukaryotic cells do not contain internal RNA polymerase III promoter sequences, they are generally not expressed in eukaryotic cells. The expression of heterologous tRNAs requires either the introduction of the sequences that direct Pol III transcription into the tRNAs or the discovery of extragenic sequences that will direct the transcription of functional tRNAs. While efforts to alter the sequences of tRNA genes to match consensus promoter sequences have not been successful, strategies for providing extragenic Pol III promoters have been successful. In yeast, we showed that a pyrrolysyl tRNA can be expressed as part of a di-cistronic construct in which an arginyl-tRNA gene provides the Pol III promoter for the transcription of pyrrolysyl tRNA, and in mammalian cells an extragenic U6 pol III promoter can be used to drive the expression of a functional orthogonal tRNA.
Genetic code expansion in animals. While it has been possible to incorporate unnatural amino acids in cells for several years, we have recently demonstrated genetic code expansion in animals. By extending the approaches we have created for controlling and imaging protein function in cells to whole animals we hope to provide new insight into processes, including embryonic development, tissue morphogenesis, tumor biology and neuronal plasticity, that can only be studied at the organism level.
We have demonstrated the incorporation of several unnatural amino acids in diverse tissues in the roundworm C. elegans. To achieve this we developed strategies for expressing a functional pyrrolysyl-tRNA synthetase/tRNACUA pair throughout C.elegans, or in specific cells, and demonstrated that this pair is orthogonal with respect to the synthetases and tRNAs of the worm. To introduce the synthetase and tRNA into worms we developed a new antibiotic transformation marker in worms. In addition we demonstrated that many unnatural amino acids can be introduced into diverse cells in the animal by simply feeding the worm bacteria grown on the amino acid.
Genetic code expansion in C.elegans: C.elegans expressing the PylRS/tRNACUA pair and a reporter containing the green fluorescent protein and mCherry separated by an amber codon. When the worms are fed unnatural amino acid substrates for PylRS they exhibit red fluorescence due to mCherry. This is imaged by fluorescence microscopy and is one of several lines of evidence for the first genetic code expansion in an animal.
In D. melanogaster we have created transgenic flies in which the pyrrolysyl-tRNA synthetase is under the control of a GAL4 upstream activating sequence and the tRNACUA is broadly expressed. We have demonstrated that this strategy allows site-specific unnatural amino acid incorporation into proteins at different developmental stages, in specific tissues and in a subset of cells within a tissue by using the wide range of GAL4 driver fly lines available. We have demonstrated that many unnatural amino acids can be introduced into the transgenic animals by simple feeding. This approach provides a foundation for probing and controlling processes in this established metazoan model organism with a new level of molecular precision.
We are currently extending the approaches we have developed in cells to map, image and control protein function in animals and provide new insight into molecular processes best studied at the organism level.
References
Proteome Labeling and Protein Identification in Specific Tissues at Specific Developmental Stages in an Animal
Nature Biotech. 2014 32:465-472.
T.S. Elliott, F.M. Townsley, A. Bianco, R.J. Ernst, A. Sachdeva, S.J. Elsaesser, L. Davis, K. Lang, R. Pisa, S. Greiss, K.S. Lilley, J.W. Chin.
[Featured in News & Views, Nature Biotech. 2014 32:445-446, Spotlight, ACS Chem. Biol. 2014 9:1068]
Efficient and Rapid C.elegans Transgenesis by Bombardment and Hygromycin B selection
PLOS ONE 2013 135:76019.
I. Radman, S. Greiss, J.W. Chin.
Expanding the Genetic Code of Drosophila melanogaster.
Nature Chem. Biol. 2012 8:748-750.
A. Bianco, F.M. Townsley, S. Greiss, K. Lang, J.W. Chin.
Expanding the Genetic Code of an Animal.
J. Am. Chem. Soc. 2011 133:1496-14199
S. Greiss & J.W. Chin.
[Featured on the BBC (www.bbc.co.uk/news/science-environment-14492948), BBC World Service Radio (http://www.bbc.co.uk/programmes/p00jttd6), Chemistry World (http://www.rsc.org/chemistryworld/News/2011/August/17081102.asp), ChemBiochem. 2012 DOI: 10.1002/cbic.201100674]
Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA synthetase/tRNA Pair.
J. Am. Chem. Soc. 2010 132:14819-14824
S. M. Hancock, R. Uprety, A. Deiters & J.W. Chin.
Genetically Encoded Photo-control of Protein Localization in Mammalian Cells.
J. Am. Chem. Soc. 2010 132: 4086-4088
A Gautier, D.P. Nguyen, H. Lusic, W. An, A. Deiters, J.W. Chin.
[Featured in Chem. Bio Chem. 2010. 11:1825-1827]
Genetically Encoding Ne-acetyllysine in Recombinant Proteins.
Nature Chem. Biol. 2008. 4: 232-234.
H. Neumann, S.Y. Peak-Chew & J.W. Chin.
[Featured in Nature 2008. 45: 868, Chemistry & Engineering News (ACS). 2008. Feb 25, p38, Chemistry World (RSC), 2008. Feb 18]