
Kiyoshi Nagai, Group Leader at the LMB and joint head of the Division of Structural Studies from 2001 to 2010, died on September 27th 2019 after a short illness.
Research begun by Kiyoshi at the LMB 30 years ago has recently culminated in a comprehensive understanding of how the spliceosome catalyses the fundamental process of gene splicing in eukaryotes. This remarkable scientific achievement is a testament to a brilliantly gifted and dedicated scientist who was an inspiration to us all.
Kiyoshi was born in Osaka, Japan on June 25th 1949. He trained as a biophysicist at Osaka University. For his PhD research in Professor Hideki Morimoto’s group at Osaka University, Kiyoshi investigated allostery in haemoglobin. During this period Kiyoshi spent 18 months at the LMB under the supervision of John Kilmartin and Max Perutz. Here he studied the oxygen binding properties of hybrid haemoglobin. After returning to Japan, Kiyoshi completed his PhD in 1977 on ‘The Allosteric Effects in Haemoglobin’. Kiyoshi then spent four years as an Assistant Professor in the Department of Physiology at Nara Medical School, Japan.
The award of a Thomas Usher Fellowship in 1981 allowed Kiyoshi to return to the LMB where he remained for the rest of his scientific career. In the early 1980s it was impossible to produce large quantity of eukaryotic proteins in E. coli. Kiyoshi worked out how to overexpress β-globin in E. coli by developing novel expression and cleavage systems. This allowed him to produce the first artificial haemoglobin mutant. In 1984 Kiyoshi was appointed as a Group Leader in the Structural Studies Division, where he embarked on the seemingly impossible task of understanding the molecular mechanism of gene splicing.
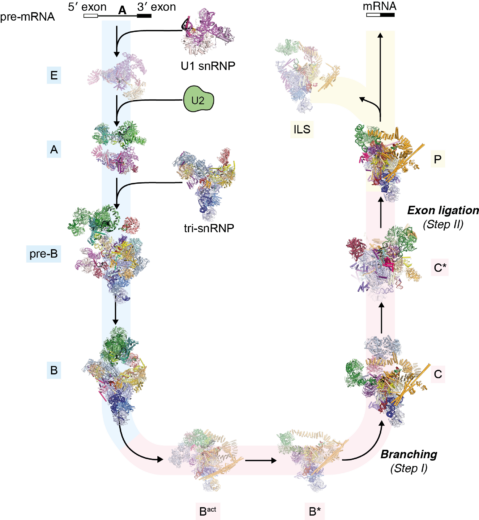
In 1977, Richard Roberts and Phillip Sharp (LMB alumni) had independently discovered that the majority of eukaryotic protein-coding genes are interrupted by non-coding segments called introns. Before the synthesized RNA transcript is decoded by the ribosome, the introns are removed and the protein coding exon segments are spliced together to generate a continuous protein coding sequence. This process is a fundamental step in gene expression. Kiyoshi pioneered structural studies of the spliceosome, the complex molecular machinery discovered by Joan Steitz and named by John Abelson (LMB alumni), that removes introns from pre-mRNA and splices together the 3′ and 5′ exon ends to produce the mature mRNA. The splicing reaction involves the assembly of the spliceosome on the pre-mRNA through the sequential interaction of multiple small nuclear ribonucleproteins particles (snRNPs) and other non-snRNP proteins (Figure: The splicing cycle reaction catalysed by the spliceosome). Because the spliceosome is a large and dynamic molecular machine, understanding the molecular basis of splicing represented a massive challenge for structural biology.
Kiyoshi first published on this topic in 1990, starting with relatively small proteins in isolation or bound to RNA, and in the process defined the first example of the widespread RNA-recognition motif domain. He steadily built up a series of RNP structures during the 1990s and early 2000s, learning how to express and assemble their components. This culminated in the spectacular crystal structure of the U1 snRNP in 2009, which gave the first glimpse of the architecture of a complete spliceosomal snRNP. This structure convincingly demonstrated that such large structures were ordered enough to crystallize, and galvanized the field – the spliceosomal structure no longer seemed quite so unattainable. Kiyoshi followed up with the structure of the U4 core snRNP in 2011, and in 2013 the crystal structure of Prp8, the central scaffold protein of the spliceosome.
The last few years have seen a revolution in structural biology as electron cryo-microscopy (cryo-EM) techniques have allowed atomic resolution structure determination without crystallization. In 2015, Kiyoshi was the first to publish a cryo-EM structure of the U4/U6.U5 tri-snRNP, providing a glimpse of the future of structural biology of in splicing. In 2016 and 2017 these were joined by structures of a spliceosome immediately after the first catalytic step of branching and the subsequent remodelled state that is competent to bind the 3’ splice site. In 2017 Kiyoshi determined the structure of the post-catalytic spliceosome, which was stalled after ligation. In this last complex, the detailed description of non-Watson-Crick base pairs between the 3’ splice site AG dinucleotide and both the branch-point adenosine and the 5’ splice site GU dinucleotide beautifully explained how the 3’ splice site is recognized. Earlier steps in spliceosome assembly were very recently resolved by Kiyoshi’s group through a structure of the pre-catalytic B complex on a pre-mRNA substrate. As a result of these structures, we can now follow an extremely intricate molecular machine as it precisely excises an intron from the pre-mRNA and ligates the two pieces to produce the mature mRNA.

It was an astonishingly rapid and complete culmination of decades of effort. These spliceosome structures, revealing the catalytic phases of the spliceosome, provided conclusive evidence showing (1) that the spliceosome is a ribozyme, (2) that the two catalytic reactions, branching and exon ligation, are catalysed by a single catalytic site, (3) how the spliceosome recognizes the beginning and the end of an intron, (4) how the spliceosome catalyses the branching and exon ligation reactions by docking and undocking substrates into the catalytic site, and (5) how this is regulated by RNA helicases.
Through this persistent and creative research effort, Kiyoshi and his team’s unravelling of the puzzle of the spliceosome, a task that only a few years ago seemed intractable, represents one of the greatest achievements in modern molecular biology.
Kiyoshi was a thoughtful, compassionate and cultured colleague, who through his concern and regard for his students, post-docs, colleagues and friends, engendered their affection and respect. Richard Henderson said: “Kiyoshi was bold, ambitious and totally fearless scientifically, as well as immensely skilled experimentally. With his warm, supportive and perceptive personality, he and his research group formed a highly successful team.” Tony Crowther commented: “As joint Head of Division with Kiyoshi between 2001-2005 I found Kiyoshi’s advice invaluable, as he had a sound judgement on scientific matters and a sympathetic and supportive approach to personal problems.”

Venki Ramakrishnan reflected that: “Kiyoshi was a real scholar, as well as a bold and creative scientist. He was also a very kind, generous and thoughtful colleague. We first became friends when I came here on sabbatical in 1991-2, found we shared many interests including RNA biology and classical music, and it is in large part due to him that I moved to the LMB seven years later. When we were Heads of the Structural Studies Division together, I was struck by how thoughtful and conscientious he was, and what an excellent mentor he was not just to his own group but to young scientists throughout the Division.”

Kiyoshi’s work was recognized by numerous awards and honours including EMBO Membership, Fellowship of the Royal Society, The Biochemical Society Novartis Prize and Medal, and Osaka University Global Alumni Fellow. Earlier this year Kiyoshi was diagnosed with inoperable liver cancer. Kiyoshi confronted this illness with the same inspirational courage and strength that he showed during his scientific career. He died peacefully on the evening of Friday 27th September in the presence of his wife Yoshiko and daughter Yuko.
Further references
Kiyoshi’s personal group page
Latest Insight on Research article