 Every minute, cells make millions of new proteins which must be transported to the correct location, folded, modified and assembled with other proteins in order to function properly. Failure at any of these maturation steps can reduce protein function and lead to the accumulation of aberrant protein intermediates, resulting in disease. To mitigate this, protein quality control factors are often directly embedded in biosynthesis pathways and act as monitors that quickly recognise and degrade aberrant proteins. Yet protein biosynthesis and quality control must be precisely balanced to first give nascent proteins an opportunity to mature and only initiate quality control processes if they fail. How cells impose a time limit on maturation processes and effectively triage nascent proteins to the appropriate fate is not understood.
Every minute, cells make millions of new proteins which must be transported to the correct location, folded, modified and assembled with other proteins in order to function properly. Failure at any of these maturation steps can reduce protein function and lead to the accumulation of aberrant protein intermediates, resulting in disease. To mitigate this, protein quality control factors are often directly embedded in biosynthesis pathways and act as monitors that quickly recognise and degrade aberrant proteins. Yet protein biosynthesis and quality control must be precisely balanced to first give nascent proteins an opportunity to mature and only initiate quality control processes if they fail. How cells impose a time limit on maturation processes and effectively triage nascent proteins to the appropriate fate is not understood.
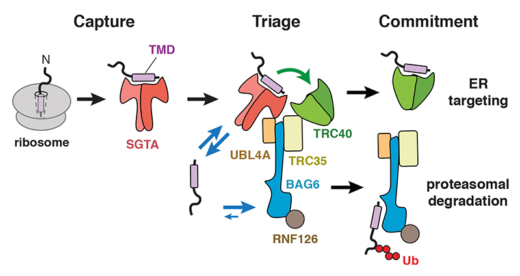
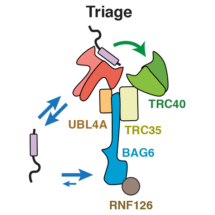
New research from Manu Hegde’s group in the LMB’s Cell Biology Division has provided insights into one such triage process. The transmembrane recognition complex comprises three chaperones (SGTA, Bag6 and TRC40) that together determine the fate of a class of membrane proteins. TRC40 is a biosynthetic factor that delivers substrates to the endoplasmic reticulum (ER) for maturation, while Bag6 is a quality control factor that facilitates substrate degradation in the cytosol. Since all three chaperones can bind the same substrates with comparable affinity, it was unclear how substrates are accurately triaged between these two very different fates.
To answer this question, Susan Shao, a former postdoc in Manu’s lab, worked with Monica Rodrigo-Brenni and Maryann Kivlen to biochemically rebuild this triage reaction in a test tube with purified proteins. To follow the reaction over time, the membrane protein substrate was selectively labeled in two ways: it was made radioactive so could be detected quantitatively and it contained a UV-activated crosslinker to ‘see’ which chaperone was holding it at any moment in time. By measuring the kinetics of substrate flux through the chaperone complex, they discovered how substrates are given a time window to preferentially target to the ER, but are then degraded if they fail. SGTA proved to be the fastest at capturing new substrates and keeps them uncommitted from any fate. Priority to ER targeting is achieved by a ‘private’ and very fast substrate transfer reaction from SGTA to TRC40, analogous to the baton transfer between two runners in a relay race. Just like the runners, if one of the chaperones is unavailable or out of position, the baton will be dropped. In this case, a monitor watching the transfer would declare the transfer a failure and clear the baton from the track. This is what Bag6 does: it picks up substrates that fail successful handover to TRC40 and dissociate from SGTA. In this way, the length of time substrates have to mature is limited by how long SGTA can hold onto them, which turns out to be around 20 seconds. Because TRC40 and Bag6 hold on to substrates 15-30 times longer than SGTA, transfer to these chaperones effectively commits substrates to ER targeting (successful completion of the race) or quality control (disqualification), respectively.
This work provides a molecular explanation and introduces new concepts for how protein triage decisions are made. This may be widely applicable to similar processes that are crucial for maintaining cell physiology and may lead to a better understanding of how protein aggregates, known to cause neurodegenerative disease, accumulate when the process goes awry.
This work was funded by the MRC, St John’s College, a Life Sciences Research Foundation fellowship, an Ellison Medical Foundation fellowship, an American Federation for Aging Research fellowship, and Harvard Medical School.