Our genetic code is translated from DNA into proteins through an intermediate molecule: messenger RNA (mRNA). One major way in which synthesis of proteins can be regulated is through turnover of mRNA; less protein is produced from a short-lived mRNA molecule. The signal for the degradation of a particular mRNA is the removal of a stretch of adenosines (As) at the end of the molecule, known as the poly(A) tail. The mRNA poly(A) tail and the deadenylation enzymes that are responsible for its removal are highly conserved across nature and the process of removal is highly regulated. However, it was not well understood how these enzymes recognised the poly(A) tail, why these enzymes degrade only the poly(A) tail and not the message itself, or why poly(A) is used rather than some other RNA sequence. Lori Passmore’s group in the LMB’s Structural Studies Division have found a very unexpected and surprising answer to these questions and identified a novel mode of single-stranded RNA recognition based on structure rather than sequence.
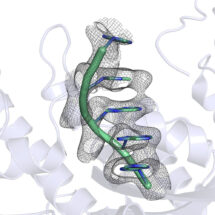
Using X-ray crystallography and biochemical assays, Terence Tang, a PhD student in Lori’s group, made the surprising finding that some deadenylase enzymes work by recognising the structure of the poly(A) tail. Enzymes that recognise RNA or DNA sequences often bind to unique features of the individual bases. However, Terence could see that, in this case, they don’t read the individual bases that make up the poly(A) tail, but instead recognise the overall shape of a series of As. This would be similar to reading text not by reading the individual letters but by recognising the overall shape of a word. It has also long been known that poly(A) RNA by itself naturally forms a helix, but this was not thought to be relevant to its function. Interestingly, Lori’s team found that the deadenylase enzymes take advantage of this helical shape and specifically recognise it, so that disruption of the helix by adding other bases inhibits removal of the poly(A) tail.
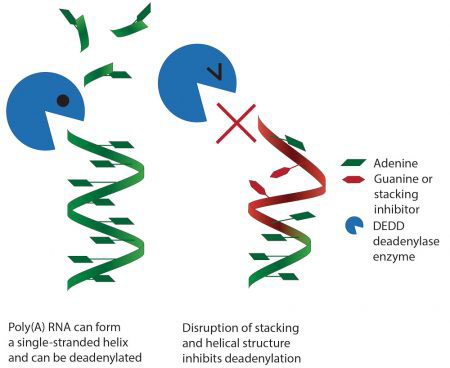
The unique ability of poly(A) RNA to form this simple helix structure and its recognition by enzymes explains why these enzymes will not remove more than the poly(A) tail and why the poly(A) tail is so highly conserved. This was a completely unexpected finding and is the first time a protein has been shown to recognise a single-stranded sequence of RNA through a simple structural organisation rather than via interactions with individual bases. Mutations in proteins involved in deadenylation have been associated with developmental defects, so understanding how these proteins work could be beneficial to our understanding of disease.
The work was funded by the MRC, ERC, and a Herchel Smith Studentship.
Further references
The intrinsic structure of poly(A) RNA determines the specificity of deadenylation. Tang, TTL., Stowell, JAW., Hill, CH., Passmore, LA. Nature Structural and Molecular Biology [Epub ahead of print]
Lori’s group page