A new technology allows parallel analysis of replication of thousands of DNA sequences in a single experiment
Nearly half of the human genome is composed of various forms of DNA repeat. Far from being merely “junk DNA”, these repeats contribute to gene function and to genome structure and evolution. One group of repeats, known as Short Tandem Repeats (STRs), have the capacity to expand at very high rates, which, if uncontrolled, could be detrimental to the organism. Pierre Murat in Julian Sale’s group, in the LMB’s PNAC Division, has revealed a mechanism that safeguards the genome from pathological expansion of STRs.
STRs, also known as microsatellites, consist of repeated motifs of 1 to 6 nucleotides placed head-to-tail and cover up to 2.5% of the human genome. Due to their repetitive nature, STRs can expand or contract during DNA replication through a process known as polymerase slippage. Although STRs share this characteristic, they differ greatly in their nucleotide composition, sequence complexity, and number of repeating units. This sequence diversity allows some STRs to fold into complex secondary structures that differ greatly from the normal DNA double helix. However, broader understanding of the evolutionary behaviour of these sequences has been limited as only a handful of the more than 5000 possible motifs has been studied.
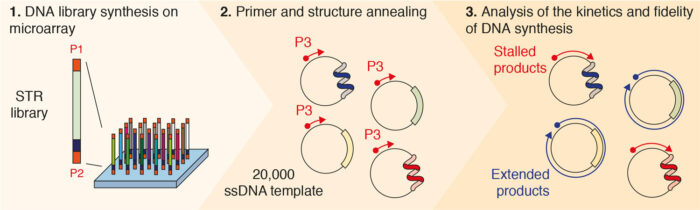
To provide a comprehensive analysis of all possible STR permutations, Pierre, working with Guillaume Guilbaud also from Julian’s group, developed a new high-throughput assay that allows measurement of DNA synthesis through thousands of sequences in parallel in a single experiment. This was then followed by deep sequencing of the replication products to assess the rate and fidelity of DNA synthesis. In total, the team monitored how well and how accurately replication was able to take place on 20,000 sequences. These correspond to all STR permutations in different lengths, as well as sequences known to impede DNA replication.
The team found that DNA polymerase stalling at STRs with the propensity to fold into complex structures triggers error-prone DNA synthesis, thereby leading to mutations that could break up the repeat and limit expansion. Indeed, correlation of their in vitro measurements of replication efficiency and fidelity with population-scale genomic data demonstrated that this behaviour of DNA polymerase at STRs, in balance with polymerase slippage, explains the complex evolutionary behaviour of these DNA repeats, including their abundance and propensity to mutate.
These findings provide a simple, and yet unexpected, model for how DNA polymerase activity at STRs dictates their stability and evolution, including a mechanism by which their expansion is constrained. Aberrant expansion of STRs to pathological lengths is the cause of several neurodegenerative diseases including Huntington’s disease, Fragile X syndrome, and amyotrophic lateral sclerosis. A better understanding of how expansion is being driven or constraints on expansion are lost might be of benefit for development of treatments for these diseases in the future.
The work was funded by UKRI MRC.
Further references
DNA polymerase stalling at structured DNA constrains the expansion of short tandem repeats. Murat, P., Guilbaud, G., Sale, JE. Genome Biology (Epub ahead of print)
Julian’s group page