The structure of the SARS-CoV-2 spike protein on the viral surface and an engineered trapped closed form of the protein could have important implications for vaccine development
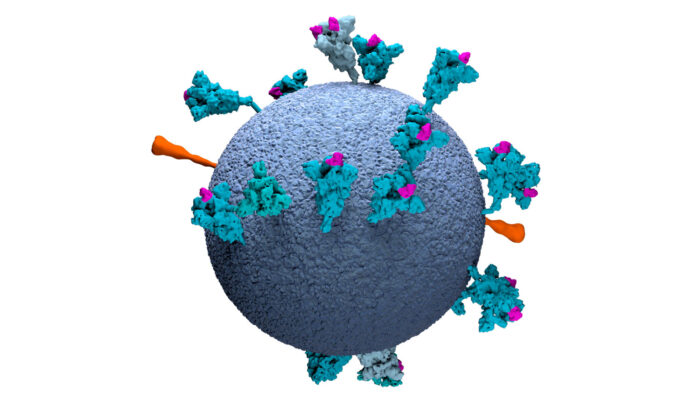
Having a detailed structural knowledge of the surface of SARS-CoV-2, the virus that causes COVID-19, will be of great benefit to both development of treatments and vaccines and development of better diagnostic tests. John Briggs’ group, in the LMB’s Structural Studies Division, has led two new studies of the spike (S) protein that protrudes from the virus surface. Spike protein attaches to the receptor on the target cell, ACE2, and is the dominant target of the immune system.
Spike proteins of SARS-CoV-2 are known to exhibit open and closed conformations. In the closed conformation the receptor binding site is hidden, whereas it is exposed in the open conformation. This has also been seen in purified SARS-CoV-2 spike proteins, but the relevance of these different conformations in the context of an intact virus has been unknown.
Using electron cryo-microscopy (cryo-EM) and electron cryo-tomography (cryo-ET), a team led by Zunlong Ke, Joaquin Oton, and Kun Qu in John’s group, and with collaborators in Sjors Scheres’ group and in Heidelberg University, determined the high-resolution structure of the spike protein directly on the surface of virus particles and describe its distribution. They found that the receptor binding domain of the spike protein is present in both open and closed conformations, showing that our immune systems will encounter the protein in both of these forms. Knowledge of how the viral surface is presented to our cells will help when developing strategies for vaccine development and improve understanding of aspects of infection and the immune response.
We do not have a vaccine for many viral diseases, due in part to the instability of their surface antigens. The instability that allows the SARS-CoV-2 spike protein to exist in different conformations might present a challenge for development of a successful vaccine against it. A team led by Xiaoli Xiong and Kun Qu in John’s group, along with collaborators at the University of Cambridge, Addenbrooke’s Hospital, Royal Papworth Hospital, Sun Yat-sen University in China, and LMB Group Leaders Andrew Carter, Sjors Scheres, and Leo James, engineered a form of the spike protein that is trapped in the closed form.
By adding some disulphide bonds that hold the spike protein in its closed formation, the team produced a form of the protein that remains intact when heated to 60ºC or when stored at 4ºC in a fridge for at least a month. One benefit of this enhanced thermostability is that it could enable better diagnostics in regions with poor infrastructure.
Stabilising the SARS-CoV-2 spike protein could be beneficial for vaccine development, as successful immunisation requires stable antigens. In addition, different conformations of the spike protein are expected to generate different antibody repertoires, and therefore provide different protection as part of a vaccine. Therefore, this closed form of the spike protein could be of use for vaccinology, as a stable antigen and to induce different immune responses from other forms of the protein.
The work was funded by UKRI MRC, ERC, Deutsche Forschungsgemeinschaft, the Japan Society for the Promotion of Science, the 100 Top Talents Program of Sun Yat-sen University, and the Wellcome Trust.
Personal perspectives of COVID-19 research
We are accompanying Insights on Research that cover COVID-19 research with interviews with the scientists who carried out the work, to give some perspective of the experiences of transitioning to COVID-19 research and working at the LMB during the height of the ongoing pandemic. Two members of John’s group who led this research on the spike protein, Xiaoli Xiong and Zunlong Ke, describe why they ended up doing this work and what it was like working on these projects.
What is your normal research focus?
Xiaoli: I had been focused on understanding influenza virus assembly before the COVID-19 pandemic.
Zunlong: Before the COVID-19 outbreak, I had been working on structural characterisation of surface glycoproteins on several enveloped viruses, including HIV-1, Ebola virus, and influenza A virus. Our goal was to understand the mechanisms of viral entry, fusion, and structural maturation.
Why did you change focus to work on COVID-linked research?
Zunlong: I was born and grew up in a village near Wuhan, China. During the four years of undergraduate study in Wuhan University, I developed my passion for the city. As a young researcher, I really hoped that I was able to contribute to the global fight against the pandemic. As it turned out, I was fortunate enough to do so and made important observations of the nature of the virus.
Xiaoli: As a high school student who experienced the SARS-CoV-1 outbreak in 2003 and someone who did research on various viruses such as influenza and SARS-CoV-1 viruses, I was particularly worried by the SARS-CoV-2 outbreak in Wuhan due to its seemingly rapid and efficient spread to other parts of the world, even after the strict Wuhan lockdown. In addition, my experience working on SARS-CoV-1 spike protein worried me about potential issues regarding coronavirus vaccine development. With the excellent research infrastructure of the LMB and support of my supervisor, I thought I could initiate this work to help solve the instability problem associated with the SARS-CoV-2 S antigen.
Why is this research important to conduct now?
Zunlong: When the lockdown happened at the end of March, the total confirmed cases worldwide was approximately 0.5 million. When we received the first sample from Heidelberg in April, the number was 1.5 million. When we submitted our manuscript at the end of June, the number of confirmed cases had gone up to 10 million! And now it is past 20 million, with more than 0.7 million deaths. There is no need to further emphasise the importance of studying the causative agent of this pandemic: SARS-CoV-2.
Xiaoli: There is currently no vaccine for COVID-19 available, and I believe antigen stability will be important for an effective vaccine. I hope the technology we developed to stabilise the S antigen will help vaccine and diagnostics development at this stage of pandemic.
What was a typical day like in the lab during lockdown?
Zunlong: Most of the time I had microscopy sessions. I would load the sample in the morning, set up automatic data screening and go home any time between 2-5 pm for lunch. After lunch, I would go back to the lab and continue the microscopy sessions. For challenging samples, I stayed overnight sometimes to either finish screening the sample so that we get the feedback or stayed up late to set up data collection.
I still remember the feeling when I walked out of the building at dawn after a day’s work, and I liked it. I felt tired but also awakened by the fresh air. It was rewarding and felt like I had achieved something, but knew there was still a lot to do.
Xiaoli: During the lockdown, I tried to advance this research as much as I could, I had been culturing bacteria for large scale DNA preps, transfecting cells, harvesting, purifying proteins and characterising them by SDS-gels and negative stain EM. If the construct looked good, cryoEM grids were prepared. There were many constructs to check and Kat and me were also trying to meet protein demand from the diagnostics labs and collaborators so it had been a laborious routine.
What has been different?
Xiaoli: The institute was dark and quiet probably because I often ended up working until rather late at night. There were very few people working in the institute and I missed finding people to discuss various topics including scientific problems.
Zunlong: During the lockdown, I was able to use any equipment whenever needed, which dramatically increased my productivity. But I missed the casual chats and coffee/tea breaks, in-person heated discussion, the scones in the morning, lunch sitting outside the canteen, and after-lunch fussball matches.
Did any action by others (apart from your co-authors of course) help or make a difference?
Zunlong: Yes, absolutely! I have had unlimited support from the EM facility scientists, including Grigory Sharov, Giuseppe Cannone, and Anna Yeates.
In addition, Darren Harper and Russell Ableman (Laboratory Services and Purchasing) really helped me out during the lockdown for shipments. There was a lot of material exchange between our collaborator and us – being able to efficiently ship materials back and forth is quite important!
Xiaoli: Besides the contributions from other co-authors, I am grateful to Grigory Sharov, Giuseppe Cannone, and Anna Yeates of EM facility and Pat Edwards of cell culture facility to keep things running smoothly. Many other people have also helped, for example, Ralf Salzer and James Stowell came to rescue when centrifuge bottles got stuck. Max Wilkinson, Danyang Zhang, and Francesca Coscia generously lent us reagents when we caught short, so big shout of “Thanks” to everyone who helped!
When did the COVID-linked research start and how long did it take to get results?
Xiaoli: We started the work in mid-March when we received the expression constructs from China, and we did not get good protein until early April, as proteins expressed from the initial constructs had stability problems. Since April we started to accumulate useful data and we submitted the paper in mid-June. This progress is definitely greatly accelerated and in normal circumstances this work would probably take more than a year to complete.
Zunlong: In February or March, I started to do literature reading on coronaviruses. One morning in the canteen, I expressed my interest to my supervisor (John Briggs) in working on this topic because it is scientifically interesting and of public importance.
We obtained our first SARS-CoV-2 sample in early April and deposited our manuscript on bioRxiv at the end of June. All the data collection was done in a span of two months. I would say this timescale is quite impressive. Normally, this project would take 1-2 years, or even longer.
What next? Will your involvement in COVID research continue or are you returning to your normal research focus?
Zunlong: The pandemic is still ongoing, and we should carry on with the basic science investigating the biology of SARS-CoV-2. There are still quite a few interesting avenues that we can follow down the path in the COVID research field, so, for now, I will continue with this work for at least another few months, or maybe a year or even longer.
Xiaoli: I came to the UK shortly after the SARS-CoV-1 outbreak in 2003 to pursue my studies in biochemistry and microbiology. After 17 years, I am heading back to China amid the SARS-CoV-2 pandemic to start up a research group in Guangzhou from September. As someone who has been long involved in structural biology of viruses, I will work on influenza virus and coronavirus, but, based on the current situation, I will be likely to focus on research that will provide support for development of a successful vaccine for SARS-CoV-2. Without a vaccine the situation may not return back to normal!
Further references
A thermostable, closed SARS-CoV-2 spike protein trimer. Xiong, X., Qu, K., Ciazynska, KA., Hosmillo, M., Carter, AP., Ebrahimi, S., Ke, Z., Scheres, SHW., Bergamaschi, L., Grice, GL., Zhang, Y., The CITIID-NIHR COVID-19 BioResource Collaboration, Nathan, JA., Baker, S., James, LC., Baxendale, HE., Goodfellow, I., Doffinger, R., Briggs, JAG. Nature Structural & Molecular Biology (Epub ahead of print)
Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Ke, Z., Oton, J., Qu, K., Cortese, M., Zila, V., McKeane, L., Nakane, T., Zivanov, J., Neufeldt, CJ., Cerikan, B., Lu, JM., Peukes, J., Xiong, X., Kräusslich, H-G., Scheres, SHW., Bartenschlager, R., Briggs, JAG. Nature (Epub ahead of print)
John’s group page
Ralf Bartenschlager (University of Heidelberg)
Hans-Georg Kräusslich (University of Heidelberg)
Ian Goodfellow (University of Cambridge Department of Pathology)
Helen Baxendale (Royal Papworth Hospital)
Steve Baker (CITIID)
James Nathan (CITIID)
LMB joins the fight against COVID-19