Development of a method that shows where proteins are bound to RNA in spliceosomal complexes provides insight into splice site proofreading mechanisms

Splicing is an essential step in the processing of pre-mRNA in eukaryotes. To do this correctly, the spliceosome must recognise and select three specific splice sites on the pre-mRNA. Studying how splicing fidelity is maintained has been difficult as some of the factors known to promote correct splice site usage bind to the spliceosome only transiently. Kiyoshi Nagai’s group in the LMB’s Structural Studies Division, in collaboration with Jernej Ule’s group at the Francis Crick Institute, have now developed a method that has allowed them to study how two such proteins bind to the pre-mRNA substrate at different stages of splicing.
Splicing proceeds through a branched RNA intermediate in which all three splice sites are aligned by the spliceosome. This alignment allows the intervening introns to be excised and exons to be joined together into the mature mRNA, which can then be used by the cell to make proteins. Prp16 and Prp22 are ATPases that use the energy of ATP hydrolysis to translocate along RNA and have been shown to proofread use of the correct splice sites as well as their proper alignment during mRNA synthesis. Previously, Kiyoshi’s group has used a mutant form of Prp22 to stall the spliceosome immediately after splicing in order to obtain its structure. Although this structure and structures of other spliceosome states revealed where these ATPases bind the spliceosome, it remained unclear how these ATPases engage spliceosomal RNAs or the pre-mRNA substrate and how they remodel the spliceosome to ensure correct splice site selection.

To address this question, Lisa Strittmatter, a former PhD student with Kiyoshi, and Charlotte Capitanchik, a PhD student with Jernej Ule, led the work to develop a method they named psiCLIP (purified spliceosome individual nucleotide resolution Cross-Linking and ImmunoPrecipitation). The technique involves first stalling and purifying the spliceosome at specific stages, to investigate how Prp16 and Prp22 engage their RNA substrate at defined points in the splicing pathway. UV is then used to cross-link RNA with any bound proteins in the spliceosome, the protein of interest is purified under stringent conditions, and the bound RNA is sequenced to determine with nucleotide resolution precisely where the protein is bound. Mapping the resulting binding profile onto cryo-EM structures of the spliceosome stalled at those same stages then allowed the team to propose a model that explains how these ATPases function.
Previous studies had proposed that Prp16 and Prp22 bind to their RNA substrate in a defined region and then remodel the spliceosome by sliding along the RNA without dissociating. Contrary to this, psiCLIP provided evidence that they bind at multiple sites on the splicing intermediate RNA and undergo multiple rounds of binding and dissociation to remodel the spliceosome. The psiCLIP profile showed that Prp22 promotes alignment of the 3’-splice site with the other splice sites by binding broadly around this region before mRNA synthesis. After splicing is completed, Prp22 binds in a more narrow region of the mRNA to promote its release from the spliceosome. Surprisingly, the team also found that Prp22 senses the stability of the spliceosome, in addition to monitoring correct recognition of the 3’-splice site by the spliceosome active site. In the absence of specific auxiliary factors that promote exon ligation, Prp22 no longer binds the RNA substrate, thus rejecting these spliceosomes and preventing erroneous exon ligation.
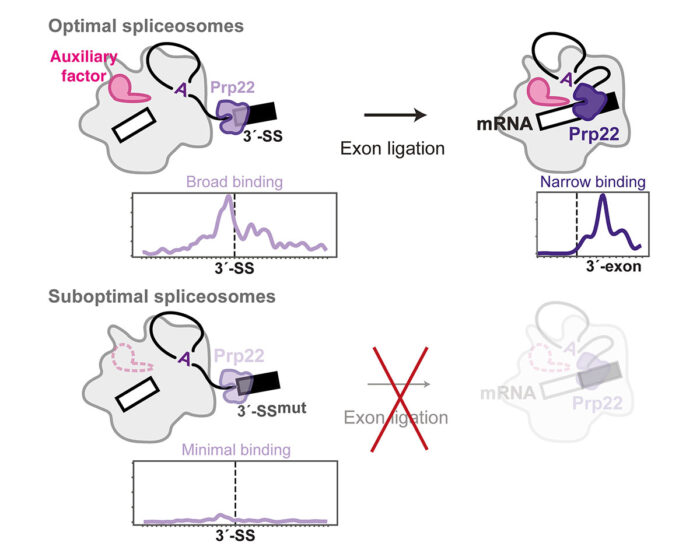
Use of the wrong splice sites can have serious consequences for alternative splicing in specific tissues. Erroneous splicing has been linked to at least 15% of human diseases, so understanding, at a molecular level, how splicing fidelity is maintained could lead to new treatments for these diseases. By preventing splicing in non-optimal conditions, Prp22’s proofreading mechanism may be particularly important in ensuring that tissue-specific auxiliary factors properly regulate alternative splicing in different parts of the body.
The work was funded by UKRI MRC, ERC, Cancer Research UK, Wellcome Trust, Boehringer Ingelheim Fonds, Winton Charitable Foundation, and the Okinawa Institute of Science & Technology Graduate University.
Further references
psiCLIP reveals dynamic RNA binding by DEAH-box helicases before and after exon ligation. Strittmatter, LM., Capitanchik, C., Newman, AJ., Hallegger, M., Norman, CM., Fica, SM., Oubridge, C., Luscombe, NM., Ule, J., Nagai, K. Nature Communications 12, 1488. https://doi.org/10.1038/s41467-021-21745-9
Kiyoshi’s group page
Jernej Ule’s group page
Previous Insights on Research
Structure of a post-catalytic human spliceosome improves understanding of splicing control
Spliceosome catalysis: the completed puzzle