Increased metabolism mediated by mTOR kinase is crucial resistance factor against TB by preventing detrimental macrophage necrosis

Although tuberculosis (TB) remains a major infectious threat, killing more than one million people each year, most infected individuals can clear the infection. The host-bacterial battle is centred in the granuloma, the hallmark complex immune structure that forms soon after TB infection. Granulomas consist of macrophages containing the infecting TB bacteria and other immune cells and are successful in eradicating infection in 90-95% of infected individuals. TB occurs in the 5-10% of cases where the granuloma is unsuccessful in achieving clearance. In these instances, the bacteria replicate within the macrophages, ultimately causing macrophage death by necrosis. The bacteria can grow in this necrotic debris less hampered by the host’s immune defences. It is from these necrotic granulomas that the bacteria can transmit best to new individuals thereby maintaining the global burden of TB. A new study from Lalita Ramakrishnan, Group Leader in the LMB’s Cell Biology Division and Head of the Molecular Immunity Unit of the Cambridge Institute of Therapeutic Immunology and Infectious Diseases (CITIID) of the University of Cambridge, has identified a major pathway of host resistance, which works by preventing granuloma necrosis caused by a specific bacterial protein.
The group identified the mTOR kinase, a master regulator of metabolism, as an early host resistance factor against macrophage necrosis. This was determined after Antonio Pagán and Lauren Lee, a postdoc and PhD student respectively in Lalita’s group, found that zebrafish mutants that did not express mTOR were hypersusceptible to TB and linked this to accelerated granuloma necrosis. They found that TB infection of macrophages rapidly causes a small increase in mitochondrial metabolism by increasing glycolysis and oxidative phosphorylation. When mTOR is lacking, this infection-induced increase does not occur and the macrophage suffers severe mitochondrial damage that in turn causes it to die by necrosis.
Lalita’s group determined that the increased mitochondrial metabolism incited by mTOR, limits a specific bacterial virulence factor, ESAT-6, from inducing mitochondrial damage. Conversely, if mitochondrial metabolism cannot be increased through the mTOR pathway, ESAT-6’s mitochondrial toxicity leads to macrophage death and the release of the mycobacteria into the forming necrotic debris that is conducive to their growth. This study has uncovered a previously unknown mitotoxic function to the mycobacterial virulence factor ESAT-6.
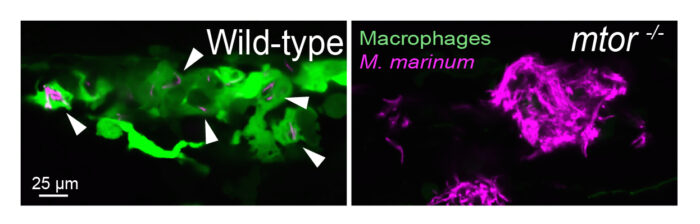
The zebrafish is an important model for the study of TB as their transparency in the early weeks of life coupled with the possibility of engineering fluorescent tags into their macrophages and other immune cells as well as into the infecting bacteria enables researchers to study the steps of TB infection in real-time. Researchers can also study the impact of introducing different drugs on the steps and outcome of TB. In this study, Lalita’s group introduced rapamycin, a human drug which inhibits mTOR, and showed that like the mTOR genetic mutation, rapamycin used at doses that only partially inhibited mTOR also caused the animals to be susceptible. Rapamycin is being considered for wide use as an anti-aging or health span promoting drug, and this study warrants caution for using it in the many areas of the world where TB is rampant.
Epidemiological analyses from Lalita and her colleagues Marcel Behr and Paul Edelstein have shown that around 90-95% of people who contract TB clear it within months, rather than harbour the bacteria for the rest of their life as has been widely thought.
This work suggests that mTOR-driven metabolism may play a major role in the clearance of TB infection by allowing infected macrophages to resist the mitochondrial toxicity of ESAT-6 so as to allow classical immune factors to come into play and kill the bacteria.
This work builds on research by David Tobin, previously a postdoc in Lalita’s group and now Associate Professor at Duke University, who first identified the mTOR mutant using mutagenised fish from an ongoing mutant screen by Cecilia Moens at Fred Hutchinson Cancer Research Center and Elisabeth Busch-Nentwich at Queen Mary University of London. Joy Edwards-Hicks and Erika Pearce, then at Max Planck Institute of Immunobiology and Epigenetics now at University of Cambridge and Johns Hopkins University, respectively, performed the metabolic profiling studies for this work.
This work was funded by the Wellcome Trust, NIH, the Max Planck Society, and the Leibniz Prize.
Further references
mTOR-regulated mitochondrial metabolism limits mycobacterium-induced cytotoxicity. Pagán, AJ., Lee, LJ., Edwards-Hicks, J., Moens, CB., Tobin, DM., Busch-Nentwich, EM., Pearce, EL., Ramakrishnan, L. Cell
Lalita’s LMB group page
Lalita’s University of Cambridge group page
How do humans resist tuberculosis? With a metabolic protein’s help – Nature Research Highlight
A deeper understanding of how tuberculosis develops – Wellcome Trust video
Previous Insight on Research articles
Pathway behind pathogenic mechanism of tuberculosis identified
New cell death pathway in tuberculosis indicates potential use of commonly used drugs