HIV-1 uses a protein net to harvest IP6 from cells and build new viruses for further infection
Animation illustrating how HIV-1 concentrates the metabolite IP6 into its virions to catalyse assembly of its iconic conical capsid.
HIV-1 is a pandemic human virus that infects 1-2 million people every year. HIV-1 evades detection by our immune system by cloaking itself in a protective shell called a capsid. In an international collaboration led by Leo James’ group in the LMB’s PNAC Division, researchers have discovered that HIV-1 builds its capsid using a metabolite called IP6 that it captures from cells with a net-like protein lattice. Disabling this capture mechanism prevents capsid construction and renders HIV-1 non-infectious.
HIV-1 replicates inside infected cells, producing thousands of new viruses which assemble and bud from the cell surface. During this process, a protein called ‘gag’ forms a hexameric lattice that coats the inside of each virus. Now, work carried out by researchers in the UK, US and Australia has revealed that this lattice creates hundreds of highly charged pockets that HIV-1 uses to harvest the metabolite IP6 from the cell. Using a method developed with Adolfo Saiardi at the Laboratory for Molecular Cell Biology (LMCB), Donna Mallery counted the IP6 molecules inside different HIV-1 viruses and found that mutants without lattice pockets lack the metabolite. Investigating the consequences of this using electron tomography, Nadine Renner discovered that IP6-deficient HIV-1 viruses fail to form a capsid properly. Even when IP6-deficient viruses manage to form a capsid, TIRF experiments performed by Anna Albecka and in Till Boecking’s lab at the University of New South Wales (UNSW) reveal they are highly unstable and collapse within seconds.
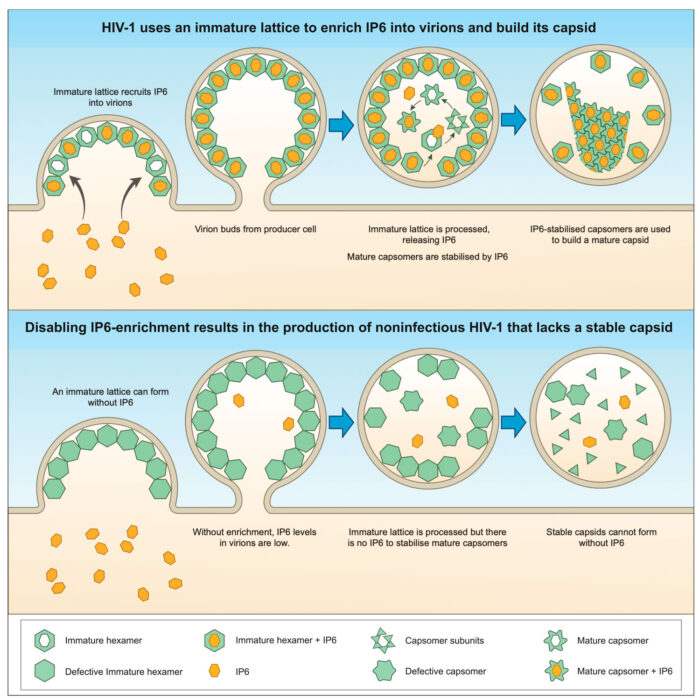
The inability to capture IP6 and form stable capsids has catastrophic consequences for HIV-1. Infection experiments carried out at LMB and by Alex Kleinpeter in Eric Freed’s lab at the National Institutes of Health (NIH) show that viruses which are unable to harvest IP6, or are produced in cells missing key IP6 biosynthetic enzymes, are almost entirely non-infectious.
The ability of HIV-1 to infect us is therefore dependent upon a small metabolite produced inside our own cells. Without it, HIV-1 cannot build its capsid or replicate. Now that we know about IP6 and the mechanism HIV-1 uses to obtain it, it may be possible to design small molecules to prevent IP6 incorporation and thus HIV-1 infection.
This work was funded by UKRI MRC, the Wellcome Trust, the NIH and the NHMRC.
Further references
HIV-1 is dependent on its immature lattice to recruit IP6 for mature capsid assembly. Renner, N., Kleinpeter, A., Mallery, D.L., Albecka, A., Faysal, K.M.R., Böcking, T., Saiardi, A., Freed, E.O., James, L.C. Nature Structural & Molecular Biology.
Leo’s group page
Adolfo Saiardi’s group page
Till Boecking’s group page
Eric Freed’s group page
Previous Insight on Research articles
Testing the capacity for intracellular antibodies to neutralise SARS-CoV-2
Understanding TRIM21 activation allows Trim-Away toolbox expansion
Furin protease is not essential for SARS-CoV-2 infection
TRIM21 links antibody and T cell immunity to combat viral infection