 New research, led by Philipp Holliger’s group in the LMB’s PNAC Division, has shed new light on the mechanism by which DNA polymerases – the enzymes responsible for replicating genomes in all animals, fungi and bacteria – are able to ensure faithful DNA replication while actively excluding damaged and/or non-cognate nucleotides from the genome.
New research, led by Philipp Holliger’s group in the LMB’s PNAC Division, has shed new light on the mechanism by which DNA polymerases – the enzymes responsible for replicating genomes in all animals, fungi and bacteria – are able to ensure faithful DNA replication while actively excluding damaged and/or non-cognate nucleotides from the genome.
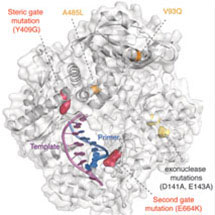
As lead researcher Christopher Cozens says: “A landmark paper by Catherine Joyce, published 15 years ago, described a general gating mechanism by which DNA polymerases exclude RNA from their active site: the “steric-gate”. However, disabling this general gating mechanism isn’t sufficient to convert a DNA polymerase into an RNA polymerase – suggesting that a second gate was in operation.”
“Our research, published in PNAS, describes a plausible candidate for the second gate, located over 25 Å from the active site in the polymerase thumb subdomain. When both the original gating mechanism and this second gate are disabled a DNA polymerase is converted to a true RNA polymerase capable of synthesizing large protein-coding messenger RNAs in minutes from RNA primers.”
The research suggests that the evolutionary path from DNA to RNA polymerases is surprisingly short, comprising just two critical mutations: the classic ‘steric-gate’ mutation in the active site (Y409G) described by Joyce and the second-gate mutation in the thumb subdomain (E664K) described by Cozens and the team. The second gate may have biological functions beyond the exclusion of RNA from the DNA genome, as disabling it also increases bypass of lesions in the DNA template.
The team’s work builds on earlier research by Vitor Pinheiro published in Science and has immediate applications in polymerase engineering and biotechnology. It is also likely to aid the study of RNA metabolism and help in the selection of aptamers – functional nucleic acids with a wide range of diagnostic, therapeutic and analytical applications – by simplifying synthesis of RNA with a modified 5′ end.
This work was supported by the Medical Research Council, European Union Grant (FP6-STREP029092 NEST), and the National Institute of Child Health and Human Development / National Institutes of Health Intramural Research Program.
Further references:
PNAS article
Holliger group page
Vitor Pinheiro in Science
Original steric-gate paper