 When the first multicellular organisms evolved, their cells needed to communicate with each other to control their growth and development. Matthew Freeman’s group from the LMB’s Cell Biology Division, led by the postdoc Markus Zettl, has shown that one way they did this was to exploit much more ancient protein quality control machinery
When the first multicellular organisms evolved, their cells needed to communicate with each other to control their growth and development. Matthew Freeman’s group from the LMB’s Cell Biology Division, led by the postdoc Markus Zettl, has shown that one way they did this was to exploit much more ancient protein quality control machinery
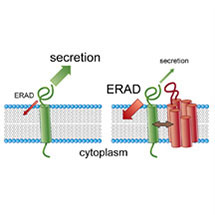
Matthew’s group has been studying the rhomboids, a family of intramembrane proteases – enzymes which cut other membrane proteins within their membrane spanning domain. They discovered a mysterious subgroup of rhomboids, which they named iRhoms, that are present in all animals (and not in single cell organisms), but lack the key catalytic signature of an active enzyme. If they are not active proteases, why are they so well conserved and what do they do?
Their work, published recently in the journal Cell, sheds light on what these “dead” iRhom proteins do. Markus Zettl and others in Matthew’s group have shown that they are located in the endoplasmic reticulum (ER), and work by binding growth factors, thereby promoting their destruction before they can be secreted as active signals. To do this they hijack a process called ERAD (ER Associated Protein Degradation), a fundamental part of the cell’s quality control machinery. ERAD normally destroys abnormal proteins, which could otherwise cause damage to the cell. In this case, ERAD destroys growth factors that are normal but not wanted at a particular stage of development. iRhoms have therefore evolved a way of exploiting the cell’s intrinsic quality control machinery to regulate growth factor signalling between cells.
There is also an interesting evolutionary angle to this story. The rhomboid proteases from which iRhoms evolved, activate the same growth factor signalling pathway that the iRhoms inhibit. So the active enzyme has been transformed into a regulator of its own signalling pathway. Matthew’s group points out that such “dead” enzymes are actually quite common. Although little is known about most of these pseudoenzymes, it is possible that many of them will turn out to have regulatory functions related to the enzymes from which they evolved.
Since signalling controls cellular behaviour – should cells divide, die, or differentiate – its disruption underlies many diseases. Rhomboids themselves have been implicated in cancer, parasitic infection, obesity, mitochondrial disorders, and others. Understanding how rhomboids are regulated, and how iRhoms control communication between cells, could provide information of potential medical significance.
Further references:
Matthew’s Group page – now at University of Oxford
Cell Paper