 The design and synthesis of genomes provides a powerful approach for understanding and engineering biology. The development of methods that can accurately replace the genome in sections, provide feedback on precisely where a given design fails and on how to repair it, and that can be rapidly repeated for whole genome replacement would accelerate our ability to understand and manipulate the information encoded in genomes. Using E. coli as a model organism, Jason Chin’s group in the LMB’s PNAC Division has made recent breakthroughs in the development of techniques to design and synthesise an organism’s genome de novo.
The design and synthesis of genomes provides a powerful approach for understanding and engineering biology. The development of methods that can accurately replace the genome in sections, provide feedback on precisely where a given design fails and on how to repair it, and that can be rapidly repeated for whole genome replacement would accelerate our ability to understand and manipulate the information encoded in genomes. Using E. coli as a model organism, Jason Chin’s group in the LMB’s PNAC Division has made recent breakthroughs in the development of techniques to design and synthesise an organism’s genome de novo.
Research by a team, including Kaihang Wang and several other members of Jason’s lab, has recently provided a system for efficient, programmable replacement of E. coli genomic DNA with very long synthetic DNA, through a technique they named REXER (replicon excision enhanced recombination). To achieve this they used CRISPR technology to cut a very long piece of synthetic DNA from a stable copy inside the cell and then used this synthetic DNA to replace a piece of the cell’s genome. The partially synthesised genome from this first step then serves as the template for the next step, leading to stepwise interchange of the original genome into a progressively more and more synthetic genome until the whole genome is replaced with a synthetic genome. The team have named this process GENESIS (genome stepwise interchange synthesis). Using these new techniques they attempted systematic recoding of the genomic sequence in an essential operon using multiple synonymous recoding schemes to remove targeted sense codons. When applied to the whole genome this may facilitate the encoded cellular synthesis of unnatural polymers by orthogonal translation systems. These results define allowed and disallowed synonymous recoding schemes, and enable the identification and repair of recoding at idiosyncratic positions in the genome.
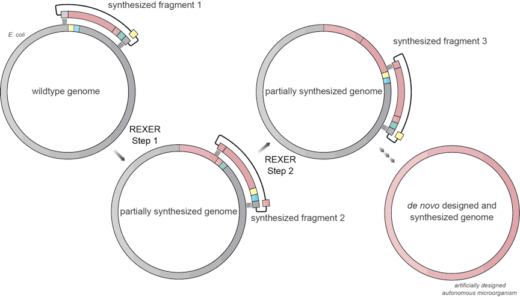 This work is potentially game changing. The development of REXER and GENESIS technologies may enable synthesis of custom-made microorganisms with artificially designed genomes. In turn, the development and validation of such genome design principles could facilitate synthesis of unnatural polymers inside living cells. Furthermore, the technologies may be used to redesign the genome of E. coli and other industry-related bacteria to potentially make pharmaceutical products, which are otherwise too expensive or impossible to produce by conventional methods. This may lead to more effective, cheaper and environmentally friendly pharmaceutical products.
This work is potentially game changing. The development of REXER and GENESIS technologies may enable synthesis of custom-made microorganisms with artificially designed genomes. In turn, the development and validation of such genome design principles could facilitate synthesis of unnatural polymers inside living cells. Furthermore, the technologies may be used to redesign the genome of E. coli and other industry-related bacteria to potentially make pharmaceutical products, which are otherwise too expensive or impossible to produce by conventional methods. This may lead to more effective, cheaper and environmentally friendly pharmaceutical products.
The work was funded by the MRC, the Danish Council for Independent Research, a Boehringer Ingelheim Fonds PhD Fellowship, the Gates-Cambridge Scholarship and by an ERC Advanced Grant.