Two proteins have been re-engineered to assemble into a flat honeycomb structure that can inhibit endocytosis of receptors from the cell surface
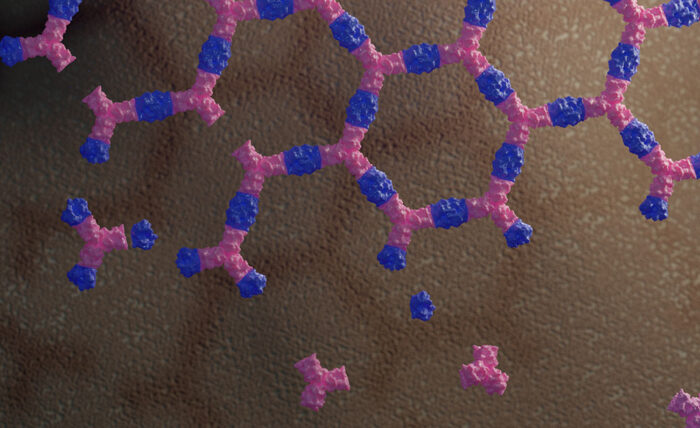
© Ian C Haydon / UW Institute for Protein Design
Proteins have evolved an enormous range of functions, from enzymes that drive reactions to cytoskeletal proteins that can remodel cell shape. The fields of artificial biology and biomaterial engineering aim to take advantage of these functions and manipulate proteins so that they have new capabilities. Emmanuel Derivery’s group, in the LMB’s Cell Biology Division, in collaboration with David Baker’s group at the University of Washington, have, for the first time, re-engineered two proteins so that they assemble into a two-dimensional material and used this design to investigate a fundamental process of cell biology: endocytosis.
One of the major aims of biomaterial engineering is to create building blocks that spontaneously assemble into materials offering desired properties at a specific time and place. This is a huge challenge to design from scratch, but if proteins can be designed so that they can form a scaffold, then adding desired properties to that scaffold by exploiting the evolved functions of existing proteins is relatively trivial.
To create such a scaffold, Ariel Ben-Sasson, in David Baker’s group, took two proteins that don’t normally interact with each other and engineered an interface between them so that their polymeric assembly results in a flat honeycomb structure. The use of two protein components allows easy control of when the material assembles as both proteins are needed. Remarkably, the actual structure formed by these proteins is almost identical to the model the team designed computationally.
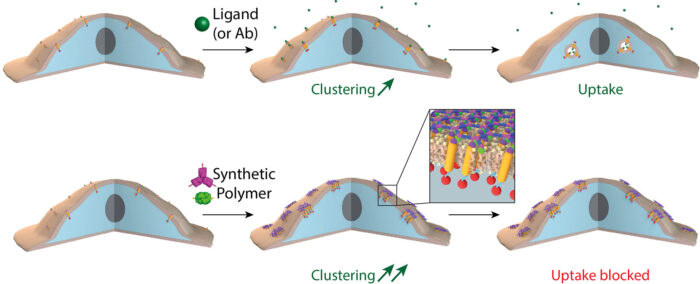
Bottom row: When receptors are clustered into small “islands” via the novel two component 2D material designed here, receptor clusters are stable at the plasma membrane for hours, or even days.
Joseph Watson, in Emmanuel’s group, reasoned that such a material could have powerful effects on cells. With simple modifications to the material, made possible by the rational protein design approach taken by Ariel, Joseph found that, when assembled on cells, the material causes cell surface receptors to very quickly cluster into “islands” under the honeycomb structure of the protein scaffold. Due to the symmetry of the 2D honeycomb lattice, the receptors are clustered in an ordered fashion. This order may, in the future, facilitate the determination of the structure of receptors by cryo-EM in situ on the cell surface.
Interestingly, if the “islands” were large enough and enough receptors were clustered together, the process of receptor endocytosis was inhibited, allowing activated receptors to stay on the surface for hours or even days. Receptor endocytosis is a fundamental process that allows cells to ensure that the response to a signal is only transient, as the receptor is internalised and degraded after activation. This new material could provide an important new tool for our understanding of the molecular mechanisms at play during this process and how we may be able to manipulate them.
The propensity of cells to internalise receptors has been proposed to lower the efficiency of immunotherapies as the antibody drugs are endocytosed by the cell along with the bound receptor, meaning that more antibody must always be injected. This new material could therefore be of benefit to immunotherapy by blocking receptor uptake.
This study is a milestone in the application of protein design to the study of cell biology, showing the potential for protein engineering to make new materials to reshape and modulate biological systems.
The work was funded by UKRI MRC, HFSP, HHMI, NIGMS, NHLBI, NCI, US Department of Energy, EPSRC, Wellcome Trust, MedImmune, Infinitus and the Max Perutz Foundation.
Further references
Design of biologically active binary protein 2D materials. Ben-Sasson, AJ., Watson, JL., Sheffler, W., Johnson, MC., Bittleston, A., Somasundaram, L., Decarreau, J., Jiao, F., Chen, J., Mela, I., Drabek, AA., Jarrett, SM., Blacklow, SC., Kaminski, CF., Hura, GL., De Yoreo, JJ., Kollman, JM., Ruohola-Baker, H., Derivery, E., Baker, D. Nature 2021
Emmanuel’s group page
David Baker’s page