 Research in the group of Kiyoshi Nagai in the LMB’s Structural Studies Division has for the first time provided detailed information on the structure and role of proteins at the active site of the spliceosome, which is responsible for the excision of introns from messenger-RNA (mRNA) precursors in the nucleus.
Research in the group of Kiyoshi Nagai in the LMB’s Structural Studies Division has for the first time provided detailed information on the structure and role of proteins at the active site of the spliceosome, which is responsible for the excision of introns from messenger-RNA (mRNA) precursors in the nucleus.
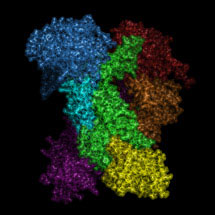
The spliceosome is a large and dynamic RNA-protein assembly. Four large RNA-protein complexes, termed U1, U2, U4/U6 and U5 snRNPs, and numerous non-snRNP proteins assemble on mRNA precursors and are remodelled extensively to form the catalytically active spliceosome. The catalytic core of the spliceosome consists of a highly structured RNA network formed between U2, U5 and U6 small nuclear RNAs (snRNAs) and conserved sequences in the pre-messenger-RNA (pre-mRNA). Andy Newman and other groups showed that Prp8, a component of the U5 snRNP crosslinks extensively with the RNA catalytic core and a pre-mRNA substrate and is therefore at the heart of the spliceosome.
In their latest paper by Galej et al., Kiyoshi’s group show that yeast Prp8 forms a cavity and that the cross-linking sites and mutations which affect splicing reactions are found in this cavity, indicating this is the site of splicing reactions.
They have also shown that Prp8 resembles proteins encoded by bacterial group II self-splicing introns in possessing both reverse transcriptase and endonuclease domains. In nuclear pre-mRNA splicing and group II intron self-splicing introns are removed by a two step reaction via a lariat intron intermediate and it was proposed that they evolved from a common ancestor. The discovery that Prp8 and group II intron encoded proteins have a similar architecture reinforces the notion that nuclear pre-mRNA splicing and bacterial group II intron splicing have a common evolutionary origin.
Retinitis Pigmentosa (RP) is a genetic disease which leads to blindness. Some cases of RP are caused by mutations in Prp8. The knowledge of Prp8 protein structure, particularly in association with its RNA partners, would provide insights into the splicing reaction. This knowledge would be useful in designing drugs to prevent splicing of disease-causing genes specifically or to facilitate splicing of genes whose splicing is prevented by mutations and potentially prevent or delay the onset of diseases such as RP.
Further references:
Article in Nature
News & Views Article
Kiyoshi Nagai’s group page