Cryo-EM analysis of structural maintenance of chromosomal complexes in E. coli reveals how two helices of DNA are simultaneously entrapped to avoid entanglements
Cryo-EM density of MukBEF bound to two segments of DNA (yellow) and the unloader MatP (blue).
Every cell in our bodies contains approximately two meters of DNA, compactly packaged into the tiny volume of the cell nucleus. To separate chromosomal regions and prevent entanglements, cells rely on structural maintenance of chromosomes (SMC) complexes. These complexes organise chromosomes in both prokaryotes and eukaryotes to enhance a wide range of DNA transactions; from transcription regulation to chromosome segregation. SMC complexes have a ring-like structure, which allows them to entrap DNA. They use their built-in motor activity to then extrude large DNA loops and change the three-dimensional path of DNA during important cellular processes.
Although SMC complexes organise chromosomes in almost all organisms on the planet, little is known about how they are able to function. Now, Jan Löwe’s group in the LMB’s Structural Studies Division, assisted by Jason Chin’s group in the PNAC Division, have determined the most complete structure of an SMC complex to date. In doing so, the research has revealed for the first time how two DNA double strands can be entrapped and encircled by the SMC protein complex.
Specifically, the team have mapped the SMC complex of E. coli, MukBEF, bound to DNA and its DNA unloading factor, MatP. Frank Bürmann, a member of Jan’s group and the study’s lead author, purified MukBEF and assembled a stable complex bound to MatP to resolve the complex structure using electron cryomicroscopy (cryo-EM). The purified MukBEF-MatP complex was shown to bind two distinct DNA helices, corresponding to the two arms of a DNA loop. The arms of the DNA loop are thread through separate compartments of the MukBEF complex, keeping the DNA loop ‘double locked’ to promote length-wise compaction of the chromosome. MatP-bound DNA threads through the MukBEF ring, whilst the second helix of DNA is clamped by the proteins MukF, MukE and the MukB ATPase heads.
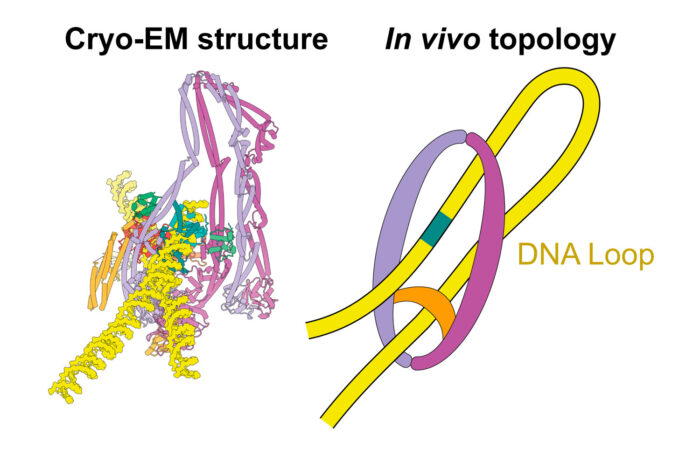
In order to test whether the mode of DNA binding revealed by cryo-EM also occurs in cells, Frank genetically engineered E. coli strains in which the SMC ring could be chemically locked. To achieve this, he teamed up with Louise Funke (a member of Jason’s group), who has developed a rapid and efficient method for genome engineering. Chromosomal DNA was indeed trapped inside the ring in these modified E. coli cells, suggesting that MukBEF does enclose DNA loops in cells – as observed in the cryo-EM structure.
This paper reveals how the SMC complex organises DNA loops to individualise chromosomes and avoid harmful tangling of DNA. This is a vital process for chromosomal organisation in almost all organisms on the planet – from bacteria to humans. The study provides an ideal basis for future investigations to better understand the structural transitions that incite SMC activities.
This work was funded by UKRI MRC and EMBO.
Further references
Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites. Bürmann, F., Funke, LFH., Chin, JW., Löwe, J., Molecular Cell
Jan’s group page
Jason’s group page
Previous Insights on Research articles
Targeting RNA replication in SARS-CoV-2
First complete atomic model of condensin lays foundation for understanding chromosome compaction