 The ability of scientists to create changes in gene sequences has improved dramatically in recent years with the emergence of a new method, dubbed ‘CRISPR’. This ‘genome editing’ technology is of great interest due to the wide range of possible applications. CRISPR is already commonly used in fundamental research to study the function of specific genes in either cultured cells or whole animal models of human biology. Potential longer-term uses of this technology include the correction of harmful mutations in humans and the destruction of viral sequences that cause persistent infections such as hepatitis and AIDS. Despite the power of current CRISPR approaches, improvements are necessary to realise the full potential of this technology. Simon Bullock’s group, in the LMB’s Cell Biology Division, has demonstrated a method that can greatly promote the efficiency of CRISPR in animal cells.
The ability of scientists to create changes in gene sequences has improved dramatically in recent years with the emergence of a new method, dubbed ‘CRISPR’. This ‘genome editing’ technology is of great interest due to the wide range of possible applications. CRISPR is already commonly used in fundamental research to study the function of specific genes in either cultured cells or whole animal models of human biology. Potential longer-term uses of this technology include the correction of harmful mutations in humans and the destruction of viral sequences that cause persistent infections such as hepatitis and AIDS. Despite the power of current CRISPR approaches, improvements are necessary to realise the full potential of this technology. Simon Bullock’s group, in the LMB’s Cell Biology Division, has demonstrated a method that can greatly promote the efficiency of CRISPR in animal cells.
CRISPR genome editing uses DNA-cutting proteins, termed RNA-guided endonucleases, that were initially discovered in bacteria. These endonucleases can be directed to cut a specific site in the genome by a short guide RNA (gRNA). When the break in the DNA is repaired it can lead to inactivation of the gene through insertion/deletion mutations or copying of a desired sequence change into the region. Many CRISPR systems currently use a single gRNA, but this can lead to inefficient gene targeting. Fillip Port, a post-doc in Simon’s group, has developed an efficient system for simultaneously expressing several gRNAs in the fruit fly, Drosophila.
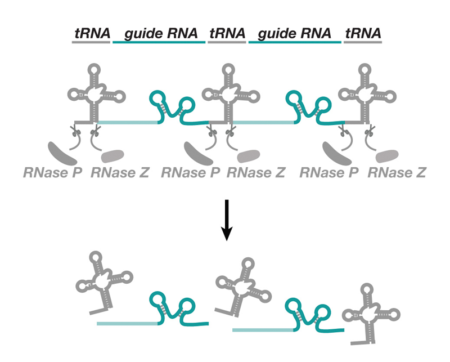
This new approach was inspired by work in rice, which had shown that multiple small RNAs could be produced from a long precursor molecule by flanking them with another type of RNA molecule, called a transfer RNA (tRNA). Because tRNA is cut in the cell’s nucleus, it can be used to liberate multiple active gRNAs for CRISPR. Fillip showed that expressing multiple gRNAs from optimised tRNA arrays greatly improves several CRISPR applications in Drosophila. Single genes can be inactivated much more frequently than with a single gRNA and multiple genes can be mutated at the same time. The study also demonstrated that the system can be adapted to inactivate genes in only certain cell types of the fly, thus allowing gene function to be studied in specific contexts.
The system is likely to be just as effective in human cells because they have very similar machinery to flies for processing tRNAs. The system may also help combat the spread of diseases such as malaria and Zika by allowing genetic modifications to be driven into wild mosquito populations. Modifying mosquito populations with a single gRNA would be difficult because variation in the genomic sequences of individuals will led to rapid emergence of genes resistant to CRISPR. However, the simultaneous use of several gRNA sequences from tRNA-gRNA arrays should alleviate this problem by hitting multiple genomic sites at the same time. The use of CRISPR technology in the laboratory is already subject to stringent risk assessment; any proposal to implement it in the wild will require further, extensive scrutiny by regulatory authorities and the public.
The work was funded by the MRC and a Marie-Curie Intra-European Fellowship.
Further references:
Paper in Nature Methods
Simon’s group page
CRISPR Fly Design