Structural study of VPS34 reveals how Rab1 and Rab5 stimulate 3-kinase activity in autophagy and endocytosis.
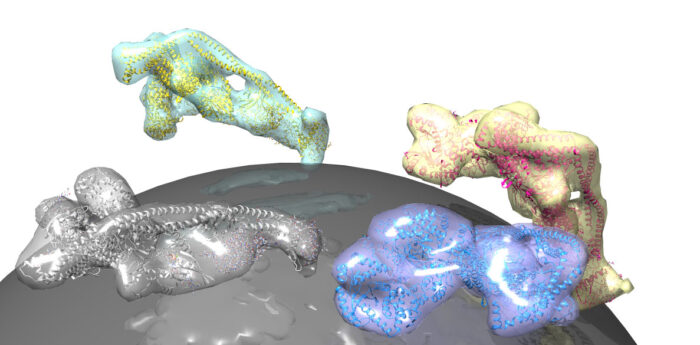
Lipid membranes envelop our cells and surround intracellular organelles and vesicles. Specific rare lipids in these membranes act as postcodes to sort these lipid compartments. One such regulatory lipid is phosphatidylinositol-3-phosphate (PI3P), which is produced by a lipid kinase named VPS34. VPS34 exists in the core of two important protein assemblies: complex I, which produces PI3P in autophagy, and complex II, which generates PI3P in early endocytic sorting. Activation of each specific process can be traced back to small, lipid-anchored G-proteins which operate like switches. Roger Williams’ group from the LMB’s PNAC Division, together with Sean Munro’s group in Cell Biology and John Briggs’ group in Structural Studies, has now elucidated how the G-protein Rab5 activates complex II in the endocytic process, and shown for the first time that the G-protein Rab1 uniquely activates complex I.
Shirley Tremel, from Roger’s group, assisted by Dustin Morado and Oleksiy Kovtun from John Briggs’ group, used electron cryo-tomography to produce a three-dimensional reconstruction of complex II on a lipid membrane. This tomographic model, which was supported by the approach of Marie-Kristin von Wrisberg in Kathrin Lang’s group at the Technical University of Munich and Zhuo A. Chen in Juri Rappsilber’s group at the Technical University of Berlin, showed that Rab5 activates complex II by nestling into a pocket on the surface of the complex. This triggers a shape change in VPS34 that releases its self-inhibition, thereby promoting production of PI3P, which facilitates sorting in the endocytic pathway.
While it has long been known that VPS34 acts as an effector of Rab5, the group sought to determine whether any other Rab GTPases directly interacts with VPS34 complexes. Jessie Bertram, from Sean Munro’s group, used proximity biotinylation to pinpoint effectors bound to mitochondrially-localised forms of 11 different human Rab GTPases. In examining all of the proteins that interact with Rab1 within cells, all four subunits that comprise VPS34 complex I were found. By reconstituting vesicles equipped with Rab1, Shirley Tremel demonstrated that Rab1 in its ‘on’ state, interacts with complex I, potently activating the complex, which may be a crucial step in triggering its autophagic role. Similar tests proved that the protein has no influence on complex II activity. Crucially, further studies led by Yohei Ohashi in Roger’s group showed that Rab1 occupies the same pocket on the surface of complex I that Rab5 occupies when binding to complex II – thus likely generating a conformational change in VPS34 to activate complex I in autophagy.
This study has answered a decades-long question as to how specific VPS34 complexes are activated. In pinpointing the conformational changes that are provoked by G proteins, the research holds significant clinical implications, as drugs that elicit these conformational changes might be useful to stimulate autophagy in therapeutic approaches to neurodegenerative diseases. Further, the tomographic methodology employed to great success here, could be applied to yield structures of other enzyme complexes that are activated on lipid membranes.
The work was funded by UKRI MRC, Cancer Research UK, European Research Council, German Research Foundation and the Wellcome Trust.
Further References
Structural basis for VPS34 kinase action by Rab1 and Rab5 on membranes. Tremel, S., Ohashi, Y., Morado, DR., Bertram, J., Perisic, O., Brandt, LTL., von Wrisberg, M-K., Chen, ZA., Maslen, SL., Kovtun, O., Skehel, M., Rappsilber, J., Lang, K., Munro, S., Briggs, JAG. Williams, RL. Nature Communications.
Roger’s group page
Sean’s group page
John’s group page