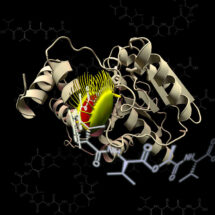
Enzymes are proteins that accelerate the conversion of substrate molecules into product molecules. Many enzymes accelerate reactions through formation of chemical bonds to their substrates, but the complexes formed this way are difficult to characterise, as they are intrinsically short-lived. In a new study, researchers from Jason Chin’s group in the LMB’s PNAC Division have developed an approach to capture a class of these enzyme-substrate complexes, known as an ‘acyl-enzyme intermediates’, that many important enzymes use to accelerate reactions. They then collaborated with Martin Schmeing’s group at McGill University and Christopher Boddy’s group at the University of Ottawa to apply the new approach to capture and characterise several key acyl-enzyme intermediates in the synthesis of valinomycin, a compound with antibiotic and anti-tumour properties. The new work provides insight into how valinomycin is made.
Acyl-enzyme intermediates are formed by the chemical reaction of particular amino acids in the active site of the enzyme with substrate molecules. By replacing the specific amino acids in the enzyme with a new non-canonical amino acid that forms chemical bonds with substrates that last many years, Nicolas Huguenin-Dezot, a researcher in Jason’s lab, was able to form very stable acyl-enzyme complexes using a model enzyme-substrate system. Diego Alonzo, a researcher in Martin Schmeing’s group at McGill University, and Nicolas then used the new approach to capture acyl-enzyme intermediates in valinomycin biosynthesis and solve the 3D structures of these intermediates using X-ray crystallography.
The incorporation of the new amino acid into enzymes, for capturing acyl-enzyme intermediates, was facilitated by genetic code expansion approaches pioneered in Jason’s lab. This approach promises to provide new insight into the acyl intermediates formed by other important enzymes. The specific insights into valinomycin biosynthesis provided by this work might suggest strategies for the creation of new bioactive compounds, antibiotics and drugs.
The work was funded by the MRC, the Canada Research Chairs Program, and the Natural Sciences and Engineering Research Council of Canada.
Further references:
Paper in Nature
Jason Chin’s group page
Martin Schmeing’s group page
Christopher Boddy’s group page
Nature News and Views: Enzymes engineered to trap reaction intermediates