The structure of the translation initiation complex provides a new model of the mechanistic processes by which the ribosome finds the start codon on an mRNA strand
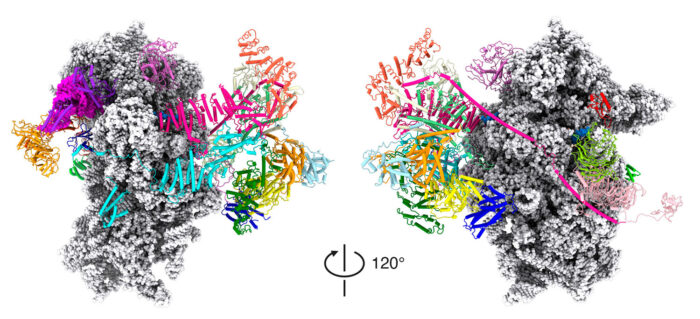
Although nearly all our cells contain our entire genome, cells use different subsets of genes to make the proteins they need to perform their various functions. This requires precise control over the processes by which the DNA is first transcribed to produce mRNA and then mRNA is translated during protein synthesis. Venki Ramakrishnan’s group, in the Structural Studies Division, has now solved the structure of the protein complex formed when mRNA is being scanned to find the start codon, providing new understanding of the molecular mechanisms underpinning initiation of translation.
Control of gene expression through regulation of transcription has been studied since the 1960s, but, over the last two decades, it has become increasingly clear that regulation of translation is equally important. Much of this control takes place at initiation of translation when the ribosome attaches to the mRNA, finds the start codon, and begins protein synthesis. There are over a dozen different proteins known as initiation factors that drive these processes, some of which are complexes almost as large as ribosomal subunits.
Recruitment of ribosomes to strands of mRNA requires the binding of a number of initiation factors to the small 40S ribosomal subunit to produce a complex known as the 43S complex, while another group of factors come together to form a complex known as eIF4F that binds to the front end of the mRNA. Although the component part of eIF4F that binds to the modified “cap” at the front end of the mRNA is known, how recruitment and then scanning are performed have been poorly understood, due to the lack of structures of the entire complex.
Structure of the human translation initiation complex
To investigate this, Jailson Brito Querido, Sebastian Kraatz and Yuliya Gordiyenko from Venki’s group collaborated with Masaaki Sokabe and Christopher Fraser at the University of California Davis, to visualise the structure of the complex with an mRNA that lacked a start codon so that it would be trapped in the act of scanning. They used electron cryo-microscopy at the LMB to obtain a structure that allowed the team to propose a model of how the mRNA slots into a channel in the small ribosomal subunit when recruited and a mechanism for how the mRNA might be pulled through the ribosome for scanning.
They were also able to predict that for most mRNAs, the start codon would need to be sufficiently far from the front end of the mRNA for it to be found in the scanning process, which was the confirmed biochemically by Sokabe and Fraser. Further confirmation of the model was obtained by mass spectrometry carried out by Mark Skehel of the LMB.
Failure to properly regulate translation, in particular during initiation, is a frequent feature of many diseases. Indeed, many of these initiation factors have been found to be dysregulated in various cancers. Knowledge of the structures of these key complexes and an associated understanding of the precise mechanisms they perform could help scientists determine how their dysregulation causes disease and develop novel treatments.
The work was funded by UKRI MRC, Federation of European Biochemical Societies, Wellcome, Louis-Jeantet Foundation, and the National Institutes of Health.
Further references
Structure of a human 48S translational initiation complex. Brito Querido, J., Sokabe, M., Kraatz, S., Gordiyenko, Y., Skehel, JM., Fraser, CS., Ramakrishnan, V. Science 369(6508): 1220-1227.
Venki’s group page
Christopher Fraser’s group page