Cryo-EM analysis of tau filament structures from a range of neurodegenerative diseases suggests a new method for characterising tauopathies
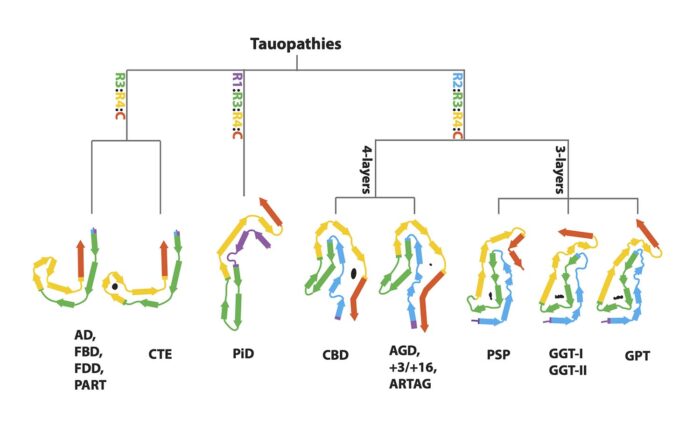
Abnormal accumulation of misfolded tau protein in filaments characterises numerous neurodegenerative diseases – collectively called tauopathies for this very reason. A long-standing collaboration between Michel Goedert’s group in the LMB’s Neurobiology Division and Sjors Scheres’ group in the LMB’s Structural Studies Division has seen the pair use cryo-EM to solve the structures of tau filaments from the tauopathies Alzheimer’s disease, primary age-related tauopathy (PART), chronic traumatic encephalopathy (CTE), Pick’s disease, and corticobasal degeneration (CBD). Now, the pair have worked again with long-term collaborators Bernardino Ghetti, from the Indiana University School of Medicine, and Masato Hasegawa, from the Tokyo Metropolitan Institute of Medical Science, to solve the structures of tau filaments from a further eight tauopathies. Their findings suggest a hierarchical classification of tauopathies, which holds important implications for future diagnostic and treatment approaches.
As with previous tauopathies, the group used brain tissues from neuropathologically confirmed cases of human diseases, extracted the tau filaments and determined their structures using electron cryo-microscopy (cryo-EM). On top of the previously studied diseases, the group have now determined the structures of tau filaments from progressive supranuclear palsy (PSP), globular glial tauopathy (GGT), argyrophilic grain disease (AGD), aging-related tau astrogliopathy (ARTAG), familial British dementia (FBD), familial Danish dementia (FDD), and inherited cases with mutations +3 and +16 in intron 10 of MAPT, the microtubule-associated protein tau gene.
Previously, tauopathies have been largely characterised through clinical diagnosis and post mortem neuropathology. When taken alongside their previous work, these cryo-EM findings suggest a new, hierarchical method by which to classify tauopathies on the basis of their filament folds. This approach has also led the group to identify a new disease, named Limbic-predominant Neuronal inclusion body 4R Tauopathy (LNT), based on the observation that the structures of tau filaments from an individual diagnosed as PSP differed from all the others.
Moreover, this method provides new ways for studying the similarities and differences between diseases. For instance, it was once thought that PSP and CBD were closely related, as they are both clinically similar 4-repeat tauopathies. However, this paper has shown that the tau folds of PSP and CBD are more disparate than was assumed. In fact, solving the structures of tau filaments from PSP has revealed a novel three-layered fold. Cryo-EM analysis has shown that PSP filaments are more similar to those of GGT, and that AGD filaments, which differ from the previous on account of their four-layered fold, are similar to those from CBD. Filaments with the AGD fold are also found in intron 10 mutation cases. Finally, the structures of tau filaments from cases of FBD and FDD are the same as those from Alzheimer’s disease and PART.
This new classification complements the previous clinical diagnostic and neuropathological approaches, and allows for the identification of new tauopathies. Even though the different folds are made of the same protein, they give rise to different diseases. The molecular mechanisms by which these different folds are formed remain unknown and are an ongoing topic of research in the groups of Michel and Sjors. Deciphering these mechanisms would hold huge implications for the diagnosis and treatment of tauopathies.
Further references
Structure-based Classification of Tauopathies. Shi, Y., Zhang, W., Yang, Y., Murzin, AG., Falcon, B., Kotecha, A., van Beers, M., Tarutani, A., Kametani, F., Garringer, HJ., Vidal, R., Hallinan, GI., Lashley T., Saito, Y., Murayama, S., Yoshida, M., Hidetomo, T., Kakita, A., Ikeuchi, T., Robinson, AC., Mann, DMA., Kovacs, GG., Revesz, T., Ghetti, B., Hasegawa, M., Goedert, M., Scheres, SHW. Nature. https://doi.org/10.1038/s41586-021-03911-7
Michel’s group page
Sjors’ group page
Bernardino Ghetti’s page
Tokyo Metropolitan Institute of Medical Science’s Dementia Research Project page
Nature News & Views
Previous Insight on Research
First structures of α-synuclein filaments from human brain
A novel tau fold in the neurodegenerative disease corticobasal degeneration
First atomic structures of tau filaments from Alzheimer’s disease
Tau filament structures differ between neurodegenerative disease
New tau structures may benefit diagnosis and treatment of head injury-associated neurodegeneration