Direct visualisation reveals FANCD2-FANCI complex is a sliding DNA clamp that monitors the integrity of the double helix but stalls at sites of single strand DNA gaps, thereby protecting damaged DNA and initiating repair, unifying its roles in crosslink repair and replication fork protection
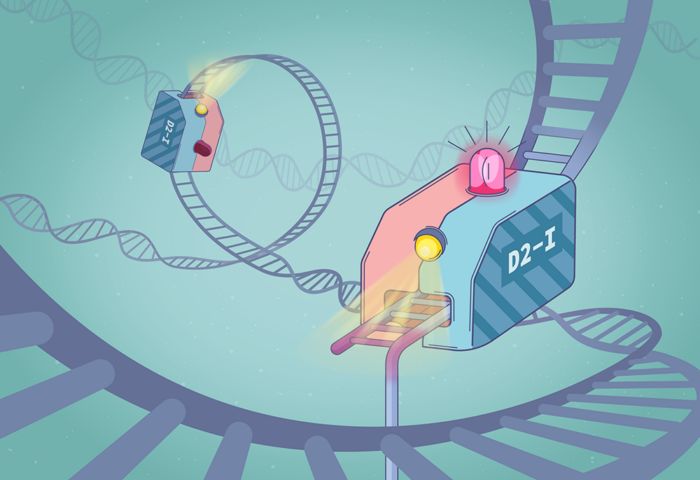
DNA is constantly damaged by radiation, chemicals, and our metabolism. Cells have developed sophisticated ways to detect and repair DNA damage, including the Fanconi Anaemia pathway which repairs DNA inter strand crosslinks – a type of damage caused by chemotherapeutics drugs and alcohol. Lori Passmore’s group, in the LMB’s Structural Studies Division, previously identified that the FANCD2-FANCI (D2-I) protein complex acts as a DNA clamp, which closes onto DNA to initiate DNA repair at crosslinks. However, key questions remained: how is the DNA damage site recognised, and why is the D2-I complex also implicated in other types of DNA damage?
Now, using a combination of single molecule-imaging and electron cryo-microscopy (cryo-EM), the Passmore group and collaborators show that D2-I identifies single strand gaps in DNA, protecting the damaged DNA and initiating repair. Such single-strand gaps in DNA appear in many types of DNA damage and at DNA replication forks, explaining the broad role of D2-I in DNA repair.
To visualise D2-I acting on DNA, Pablo Alcón, a member of Lori’s group, worked with Artur Kaczmarczyk and Korak Ray in David Rueda’s group at the MRC Laboratory of Medical Sciences (LMS) and Imperial College London. They used optical tweezers to catch a single DNA molecule between two beads, which allowed them to precisely manipulate the DNA and incubate it with chosen proteins. Using fluorescently labelled D2-I and single-molecule imaging, they observed how D2-I binds to and slides along DNA, scanning the double helix. They found that single-stranded DNA gaps, a specific type of DNA lesion caused by the breakage of one DNA strand, causes the sliding D2-I to stall at the junction between the double-stranded and single-stranded DNA.
To visualise what was happening in close detail, the team used cryo-EM to determine the structures of D2-I both in its sliding position and stalled at single-stranded/double-stranded DNA junctions. This revealed that recognition of this junction is distinct from recognition of double-stranded DNA. Cryo-EM also allowed the team to identify the KR helix – a specific region of the FANCD2 protein which is critical for recognising and stalling at these DNA legions. Disrupting the KR helix resulted in impairments to DNA sliding in vitro. Working with Guillaume Guilbaud and Julian Sale in the LMB’s PNAC Division, and Themis Liolios and Puck Knipscheer at the Hubrecht Institute, Netherlands, the group showed that mutations to the KR helix dramatically affected DNA repair in cells and Xenopus egg extracts.
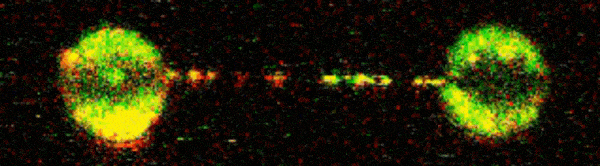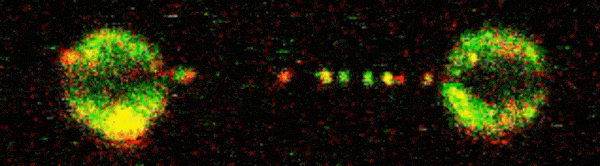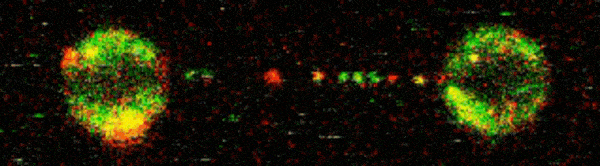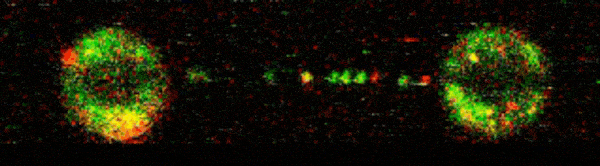
This research outlines the mechanism whereby D2-I monitors the integrity of double-stranded DNA by scanning its length and clamping down when it encounters a single-stranded junction. This flags the damage to repair enzymes and also prevents further damage by anchoring the remaining part of the double-stranded DNA in place. Understanding the process of DNA repair, and critically why it fails, holds huge importance as DNA damage is a key factor in several diseases. Insights into the molecular basis of DNA repair may help researchers to better understand these diseases, and ultimately could contribute to new therapeutic interventions or preventatives.
This work was funded by UKRI MRC, the Wellcome Trust, the European Research Council and EMBO.
Further references
FANCD2-FANCI surveys DNA and recognizes double to single-stranded junctions. Alcón, P., Kaczmarczyk, A.P., Ray, K.K., Liolios, T., Guilbaud, G., Sijacki, T., Shen, Y., McLaughlin, S.H., Sale, J.E., Knipscheer, P., Rueda, D.S., Passmore, L.A. Nature
Lori’s group page
David Rueda – MRC LMS
David Rueda – Imperial College London
Puck Knipscheer – Hubrecht Institute
Previous Insight on Research articles
How is the Fanconi Anaemia pathway activated to remove DNA lesions?
Proteins required for processing the end of mRNAs identified