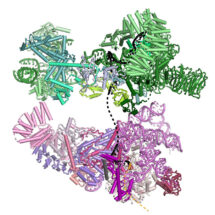
Kiyoshi Nagai’s group in the LMB’s Structural Studies Division have used electron cryo-microscopy to solve the structure of the prespliceosome at near-atomic resolution, providing new insights into how the spliceosome is assembled and regulated.
The human genome contains approximately 20,000 genes, which when transcribed produce precursor messenger RNA (pre-mRNA) consisting of coding sequences (exons) and non-coding sequences (introns). The spliceosome is the molecular machine which cuts out the introns and stitches the useful exons together, so that protein can be produced from the resulting mature mRNA. Importantly, many human genes can undergo alternative splicing: some exons can be skipped to produce variant forms of the resulting protein. This helps regulate gene expression and increases the repertoire of proteins that can be made by the cell.
Previous research from Kiyoshi’s group revealed structural details of how introns are removed in a two-step process. First, the start of an intron is recognised, cut, and joined to a specific point in the middle of that intron, forming a lasso-like looped structure. In the second step, the end of the intron is recognised, cut, and the exons joined together. It is also known that different alternative splicing factors associate with the spliceosome at an early stage in the splicing process, however the structural basis of early events in spliceosome assembly have remained poorly understood. New work from Kiyoshi’s group has now used electron cryo-microscopy to uncover the structure of the prespliceosome at near-atomic resolution, revealing that the formation of the prespliceosome is a critical step in determining which protein variants are produced during splicing.
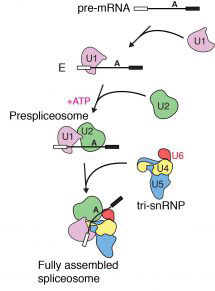
Clemens Plaschka, Pei-Chun Lin and Clément Charenton from Kiyoshi’s group made a yeast cell extract which could perform splicing. They then added a particular form of pre-mRNA which stalled the splicing reaction after prespliceosome formation so that lots of prespliceosome accumulated. Using electron cryo-microscopy they were able to collect thousands of images of the prespliceosome in different orientations which when combined together gave them a high resolution 3D image of the prespliceosome. The structure of the prespliceosome revealed how this molecular machine decides where the boundary between the protein coding exons and non-coding introns are. In addition, it provides the first insight into how human alternative splicing factors increase the use of weak splice sites.
Many human diseases, such as thalassemia, are caused when splice sites are mutated and so are no longer recognised correctly by the splicing machinery. Other diseases, including tauopathy, are caused by an imbalance of different protein variants in the cell produced by alternative splicing. Better understanding of the mechanism of alternative splicing will lead to improved insights into the molecular pathology of these conditions.
This work was funded by the MRC, the European Research Council and an EMBO Fellowship.
Further references:
Paper in Nature
Kiyoshi’s group page
Previous Insight on Research: Spliceosome catalysis – the completed puzzle