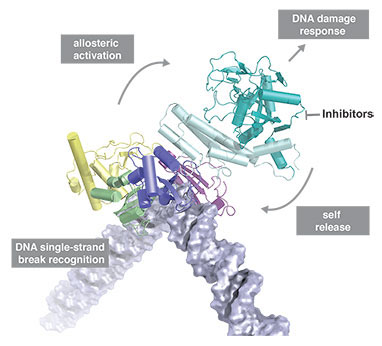
DNA damage represents a constant threat to the integrity of genomic information in cells and is closely linked to the origins of cancer. DNA single-strand breaks (SSBs) are the most frequent type of damage with thousands of such lesions from different sources occurring in each cell every day. Despite many advances in understanding the mechanistic basis of DNA repair, in the case of SSBs it has remained unclear at a detailed structural level how cells detect and signal their presence to the DNA repair machinery. Although it has long been known that it is the chromatin-associated protein PARP-1 that carries out these functions in most eukaryotes, the inner workings of this enzyme have remained an unsolved puzzle, evading conventional structural approaches and leaving the mechanism by which it acts as a “molecular nick-sensor” largely unknown until now.
Recent research led by David Neuhaus and Sebastian Eustermann in the LMB’s Structural Studies Division, in collaboration with John Pascal’s group at Thomas Jefferson University (now at Université de Montréal), has for the first time established the structural basis for DNA single-strand break recognition by PARP-1. The team found that the key to understanding how PARP-1 works is the same feature that made previous approaches so difficult, namely the inherent flexibility both of the enzyme and of a double-helix with a broken strand. While in its free state PARP-1 behaves like “beads on a string” comprising six flexibly-linked domains, in the presence of a DNA single-strand break the situation changes completely as the domains start to cooperate. By using its two N-terminal zinc fingers, PARP-1 senses the deformability of an SSB, bending and twisting the DNA at the break into a shape that undamaged DNA could never reach. This process ensures specificity and triggers a cascade of further dynamic interactions within the remaining part of PARP-1. In effect, this transfers energy from the N-terminal DNA binding interaction through the enzyme to the C-terminal catalytic domain, triggering a burst of poly(ADP-ribose) synthesis that provides the signal that damage has been found (an accompanying paper from the Pascal group and collaborators, in part building on these results with SSBs, describes changes in the catalytic domain resulting from the domain assembly process). Modification of a large number of proteins with poly(ADP-ribose) is one of the earliest signals in response to genotoxic stress in eukaryotes, and brings about concerted changes in chromatin structure as well as recruitment of DNA repair proteins.
The central importance of dynamics in this mechanism makes NMR spectroscopy a particularly powerful method for studying how PARP-1 and SSBs interact. By combining key information from NMR experiments on SSB complexes in solution with crystallographic results from the Pascal group obtained from (less flexible) complexes with DNA duplexes, the team were able to solve this previously intractable problem; integrating the two techniques in this way uses the strengths of each to overcome limitations of the other, and is likely to be an approach that is increasingly used in the future. The unusual behaviour of PARP-1 in response to DNA single-strand breaks that this study revealed could have parallels for proteins in other signalling pathways, and the results might suggest how PARP-1 looks when it is trapped on such breaks by inhibitors.
The way in which DNA damage is detected by the cell is of fundamental importance, particularly because it is highly relevant to the origins of cancer. PARP inhibitors are becoming increasingly important in cancer treatment; normal cells can tolerate the loss of PARP-mediated DNA repair caused by PARP inhibitors, but cancer cells that intrinsically lack a second repair pathway due to a genetic defect are selectively killed due to the cytotoxic effects of lacking both repair pathways. Such cumulative effects are called “synthetic lethalities”, and PARP inhibitors are the first anti-cancer drugs to exploit this idea so as to target tumour cells selectively. A better understanding of how PARP-1 works, i.e. how it recognises and is activated by SSBs, could help with the design of improved therapeutics for cancer treatment in the future.
This work was funded by the MRC, Worldwide Cancer Research, Boehringer Ingelheim Fonds, EMBO, Sidney Kimmel Cancer Center and the NIH.