 Effective immunity to infections requires the development of a diverse repertoire of antibodies. Antibody diversity is created through a process known as somatic hypermutation, which is the programmed mutation of specific sequences of DNA in the antibody genes. The introduction of mutations results in the production of antibodies that recognise and bind to different antigens, such as microbes, and allows the immune system to adapt as it is exposed to new antigens. For the first time, Julian Sale’s group in the LMB’s PNAC Division, have shown that the presence of the non-standard histone H3.3 in the chromatin of antibody genes promotes the process of antibody diversification.
Effective immunity to infections requires the development of a diverse repertoire of antibodies. Antibody diversity is created through a process known as somatic hypermutation, which is the programmed mutation of specific sequences of DNA in the antibody genes. The introduction of mutations results in the production of antibodies that recognise and bind to different antigens, such as microbes, and allows the immune system to adapt as it is exposed to new antigens. For the first time, Julian Sale’s group in the LMB’s PNAC Division, have shown that the presence of the non-standard histone H3.3 in the chromatin of antibody genes promotes the process of antibody diversification.
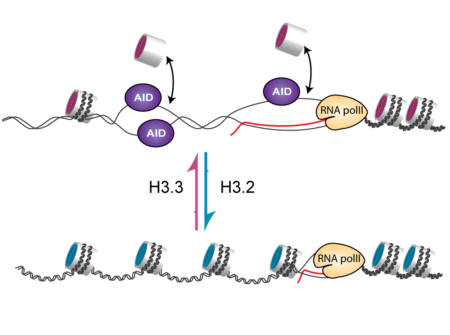
The process of hypermutation is driven by an enzyme called activation induced deaminase (AID). AID deaminates deoxycytidine (dC) bases in the DNA of the antibody genes creating deoxyuracil (dU). The recognition and processing of dU results in mutations that generate antibody sequence diversity. However, AID can only work on single stranded DNA, and within the nucleus DNA is packaged in a protein/double stranded DNA structure known as chromatin. In order for single stranded DNA to be exposed, the structure of chromatin must be loosened. At the heart of chromatin are nucleosomes, which are made up of DNA wrapped around histone protein molecules. Different components and protein modifications of nucleosomes are known to be associated with tightly or loosely packed chromatin, but the chromatin factors that promote formation of single stranded DNA during the process of hypermutation have not been fully understood.
H3.3 is a histone variant that can replace the standard H3 in the nucleosome. The effect of H3.3 on nucleosome function is still poorly understood, but it is known to be enriched in actively transcribed genes. By comparing normal cells and those that lacked H3.3, the Sale group showed that the presence of H3.3 in the chromatin of the antibody gene is necessary for efficient access of AID to the DNA and hence for antibody diversification. Surprisingly, this happens without any detectable effect on the transcription of the gene itself. Further investigation revealed that the presence of H3.3 increases antibody diversification by making the DNA of the antibody gene more likely to become single stranded. Although the exact reason for this remains to be further explored, it suggests that nucleosomes containing H3.3 are more readily displaced from the DNA during transcription allowing the negative supercoiling generated by transcription to cause short ‘denaturation bubbles’ in which the two strands of DNA are transiently separated. These short single stranded tracts provide an ideal substrate for AID to deaminate dC, thereby driving antibody diversification. This provides an important piece in the jigsaw of understanding how a diverse antibody repertoire is generated, as well as shedding light on the effects of H3.3 on chromatin function.
This work was funded by the MRC, the Darwin Trust of Edinburgh and EMBO.