
Scientists in Lori Passmore’s group in the LMB’s Structural Studies Division have revealed new mechanistic insights into the link between translation and mRNA decay. In collaboration with Jeff Coller’s group at Case Western Reserve University, USA, and Brenton Graveley’s group at the University of Connecticut, USA, the scientists’ findings have implications for understanding regulation of gene expression in eukaryotes and provide insights into an important problem which had remained difficult to address for the past two decades.
In every cell, specific parts of DNA – genes – are transcribed into mRNA, which is then translated to produce proteins, the molecular machines of the cell. mRNAs can either exist transiently or last for a long time within the cell, and these differing lifetimes are important for regulating gene expression – the longer an mRNA is present, the more of its corresponding protein will be made. The lifetime of an mRNA is influenced by its poly(A) tail – a string of bases which are added to one end of the mRNA after it is initially made. To initiate mRNA decay, this protective poly(A) tail must be removed – this is accomplished by the molecular machine Ccr4-Not, a multiprotein complex. Recent work has shown that translation rates play a major role in establishing mRNA decay rates. However, the mechanistic details of how translation is coupled to mRNA decay have been unclear.
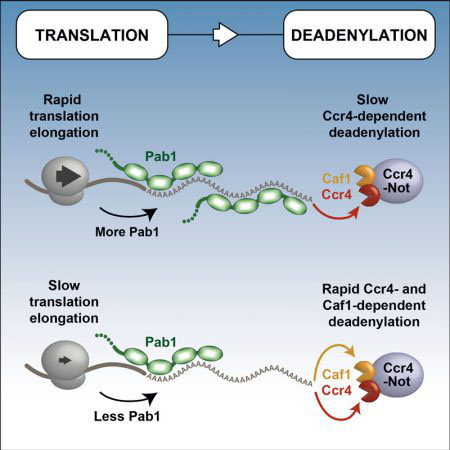
Michael Webster, who drove the project, and James Stowell – both scientists in Lori’s group – purified the seven subunit Ccr4-Not complex, accessory proteins, and model mRNAs. They then added them all to a test tube where they assembled into the complete active complex. Michael was then able to study the activity and regulation of Ccr4-Not in this test tube set-up. He showed that the Ccr4-Not complex is able to shorten poly(A) tails, even when the tails are bound by a special protein, Pab1. This was surprising as Pab1 was previously thought to protect the poly(A) tail from Ccr4-Not. In addition, the scientists discovered that the two nucleases in the Ccr4-Not complex have different functions – one nuclease (Ccr4) is able to shorten poly(A) tails bound by Pab1, whereas the other (Caf1) only trims naked poly(A) tails.
Lori’s collaborators in the USA then extended the findings into living yeast cells, making mutations in Ccr4-Not and testing the effect on mRNA stability and poly(A) tail length. This revealed that Ccr4 is a general deadenylase which acts on all mRNAs. In contrast, Caf1 is specialised and is required for the rapid decay of mRNAs which are translated slowly and have less Pab1 bound to their poly(A) tails. This therefore explains how Ccr4-Not links translation and mRNA decay rates.
This work provides new insights into the interplay of the many molecular machines inside our cells that work together to control gene expression. By better understanding the fundamental mechanisms of gene expression, Lori’s group and collaborators will also gain insights into how this process is deregulated in diseases, including developmental disorders and cancer.
This work was funded by the MRC, European Research Council and NIH.
Further references:
Paper in Molecular Cell
Lori’s group page
Jeff Coller’s group page
Brenton Graveley’s group page