A collaborative team from the LMB’s Cell Biology and Structural Studies Divisions has identified a cellular factor that detects ribosome collisions. The ribosome is the molecular machine responsible for reading the genetic code to produce proteins, a process known as translation. Such collisions between ribosomes are a sign that something has gone awry during translation, and the collision-detecting factor is critical for initiating pathways to resolve the problem. It is known that if stalled ribosomes are not resolved efficiently in mice, they develop neurodegeneration. Thus, the new research sheds light on how our body monitors its protein production assembly lines so problems can be promptly resolved before they cause cellular stress and disease.
Each cell in our body contains millions of ribosomes that translate messenger RNAs (mRNAs), the carriers of the genetic code, into proteins, the factors that carry out most of the biochemical reactions essential for life. In highly active cells, most mRNAs have between 4 to 12 ribosomes along their length, each translating around 6 codons per second. A healthy cell relies on the smooth operation of these protein-producing assembly lines. Excessively slow ribosomes are one indicator that something may not be working correctly, so cells have a process called ribosome-associated quality control (RQC) to resolve the issue. The RQC pathway recycles problematic ribosomes and initiates degradation of their associated mRNA and partially synthesized protein. But how do cells selectively detect slow or stopped ribosomes?
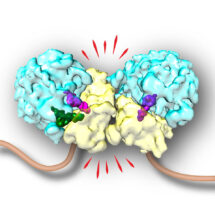
In earlier work, Szymon Juszkiewicz from Manu Hegde’s group in the LMB’s Cell Biology Division had discovered that a critical factor in the RQC pathway is a protein called ZNF598. This protein attaches a ubiquitin tag to a specific site on the ribosome, which turned out to be required to initiate the RQC pathway. In a new study, Szymon investigated what ZNF598 actually recognises, expecting it to bind to stalled but not actively translating ribosomes. Unexpectedly, he found that ZNF598 only bound to a stalled ribosome when another ribosome had collided into the back of it.
In parallel, Vish Chandrasekaran from Venki Ramakrishnan’s lab in the LMB’s Structural Studies Division was investigating poly-ribosomes, suspecting that their distinctive architecture might be recognised by the cell in some circumstances. Vish worked out methods to isolate such collided poly-ribosomes and determined their structure using cryo-electron microscopy. The structure showed that two collided ribosomes are in a precise orientation such that the ubiquitin tag attachment site for ZNF598 is near the interface between ribosomes. By recognizing this distinctive interface, ZNF598 is able to detect multiple causes of translation slowing while avoiding all normally translating ribosomes.
Understanding how cells detect faulty translation is important because the accumulation of defective proteins is a major cause of disease in humans, particularly neurodegeneration. The team is now trying to understand precisely how ZNF598 interacts with collided ribosomes, how the ubiquitin tag attached on collided ribosomes is used for their disassembly, and which cellular mRNAs are particularly prone to problems that induce ribosome collisions.
This work was supported by the MRC, the Wellcome Trust, the Agouron Institute, and the Louis-Jeantet Foundation.
Further references:
Paper in Molecular Cell
Manu Hegde’s group page
Venki Ramakrishnan’s group page