Multidisciplinary techniques reveal the structures and subunit assembly of GluA4 AMPARs in the mammalian cerebellum that support cell-type specific functions
AMPA receptors (AMPARs) are key membrane proteins which act as gates within the nervous system. They mediate fast transmissions of signals between synapses, which is crucial to memory formation and learning. Throughout the brain, AMPARs are hugely diverse due to their combinatorial assembly from four subunits, GluA1-4, auxiliary proteins such as the Transmembrane AMPAR Regulatory Proteins (TARP) family and post-transcriptional modifications. AMPAR diversity is exemplified in the cerebellum, where GluA4 subunits are uniquely enriched. Now, Ingo Greger’s group, in the LMB’s Neurobiology Division, have determined the structure and organisation of GluA4-containing AMPARs and their associated TARP subunits from the mammalian cerebellum.
The group utilised a multi-disciplinary approach to study these receptors in their natural environment, beginning by using a GluA4-specific nanobody to isolate GluA4 subtypes from pig cerebella. Mass spectrometry analyses revealed that GluA4 AMPARs are predominantly enriched with GluA1 subunits and with TARPs, with other auxiliary subunits associating at substantially lower levels.
To further probe GluA4-containing receptor populations, group members Alex Scrutton and Nayanika Sengupta conducted electron cryo-microscopy (cryo-EM) of receptors isolated from pig cerebellum, using a nanobody developed and isolated by Veronica Chang and Kuni Suzuki from the Aricescu group. They discovered that cerebellar GluA4 AMPARs segregate into two distinct sub-types, a hexameric form associated with two TARPs which largely lacked GluA2 subunits, and an octameric complex with four TARPs that incorporated GluA2. As the inclusion of GluA2 subunits results in calcium impermeability, this revealed that the GluA1/A4 population of AMPAR receptors is calcium-permeable and originates from glial cells, whilst the mixed GluA2/A4 population is calcium-impermeable and predominantly of neuronal origin.
To gain a deeper understanding of the subunit composition and gating properties of the different subtypes, group member Josip Ivica conducted electrophysiological recordings from defined receptor combinations reconstituted into HEK293 (human embryonic kidney) cells. This was supported by Imogen Stockwell, who carried out recordings from glia within cerebellar brain slices, and from cultured hippocampal slices, transfected with GluA4 mutants. These techniques exposed how the two identified receptor classes also contrast in their associated TARPs, which each have differing functional properties; hexamers associate with Type-II TARPs (mostly TARP- γ7), whilst octameric formations contain Type-I TARPs.
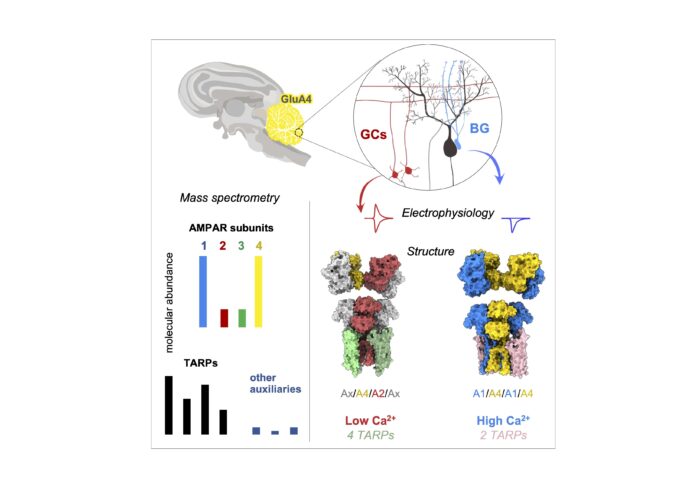
This study reveals the various stoichiometries and organisation principles of GluA4 AMPARs and their associated subunits in the cerebellum, shedding new light on how different AMPAR subtypes work together to support specific cellular functions in the cerebellar circuitry. The results highlight that both neurons and glial cells have unique receptor assemblies supporting their highly diverse signalling functions. The finding that cerebellar receptors come in distinct and diverging forms, with clear differences in calcium permeability and auxiliary subunit partners, underlines the elegant molecular diversity at the core of cerebellar function. Looking at the wider picture, this research ultimately advances our understanding of how the brain finely tunes its communication systems to support precise motor control and learning.
This work was funded by UKRI MRC and the Wellcome Trust.
Further references
Structure and organization of AMPA receptor-TARP complexes in the mammalian cerebellum. Scrutton, A.M., Sengupta, N., Ivica, J., Stockwell, I., Peak-Chew, S., Singh, B., Suzuki, K., Chang, V.T., McLaughlin, S.H., Krieger, J.M., Aricescu, A.R. and Greger, I.H. Science
Previous Insight on Research articles
Architecture of the disease-prone GluA3 receptor unlocks new avenues for drug design
How protons tune AMPA receptor-mediated information processing at neuronal synapses
First Cryo-EM structures of homomeric GluA1 AMPA glutamate receptor reveals functional roles for N-terminal domains