Studies of mycobacteria from cystic fibrosis patients reveal key steps in how they evolve to become virulent amongst humans
Mycobacteria typically live in soil and water, but pose the threat of evolving to cause transmissible human infection – the most well-known example of this is Mycobacterium tuberculosis. Recently, a species named Mycobacterium abscessus (M. abscessus) has emerged as a considerable threat to people with cystic fibrosis (CF) and other chronic lung conditions, where it causes accelerated lung damage, is often incurable and regularly prevents safe lung transplantation. Andres Floto’s group from the Molecular Immunity Unit, which is housed at the LMB have studied the evolution of M. abscessus in collaboration with Julian Parkhill’s lab at the Department of Veterinary Medicine, University of Cambridge. Their work has led to a generalisable model of mycobacterial pathogenic evolution.
The first evolutionary step the team identified is horizontal gene transfer to some environmental clones of M. abscessus, causing a large single-step change in their behaviour. Analysis of the total gene content across the species, known as the pangenome, allowed the team to identify genes acquired by the new virulent clones of M. abscessus, including a novel DNA methylase, which enhanced the bacteria’s ability to survive within human cells. The team also noted that similar horizontal gene transfer occurred in the early stages of evolution of other mycobacteria into human pathogens, thereby suggesting that this step is an important general mechanism in facilitating the behavioural changes needed for a bacterium to become pathogenic.
Secondly, the group demonstrated how, over the course of an infection, the bacteria adapts within its host – specifically in the case of M. abscessus within the lungs. The highly compartmentalised anatomy of the lung enhances the evolutionary development of the bacteria, by allowing parallel evolution of M. abscessus in different parts of the lung. Furthermore, as infections amongst CF patients are often chronic, this provides the bacteria years to cultivate their ability to thrive in human cells. Josie Bryant, a Henry Wellcome Fellow in Andres’ group, was able to identify bacterial genes which frequently mutated within patients, and showed how these alterations led to improvements in the survival ability of the bacteria.
In order for within-host adaptation to be sustained amongst the bacteria’s population, it requires transmission of these more virulent bacteria between people. Importantly, the team discovered that adapted strains of M. abscessus had a consistently low transmission rate as they proved unable to survive on inanimate surfaces, therefore limiting their ability to spread freely. Contrastingly, strains that had yet to go through the second evolutionary stage of within-host adaptation had much higher survival rates on these surfaces. This is significant as it is currently thought that most transmission of the bacteria is indirect via environmental intermediates, thus restricting the potential for M. abscessus to evolve into an full-time, obligate pathogen.
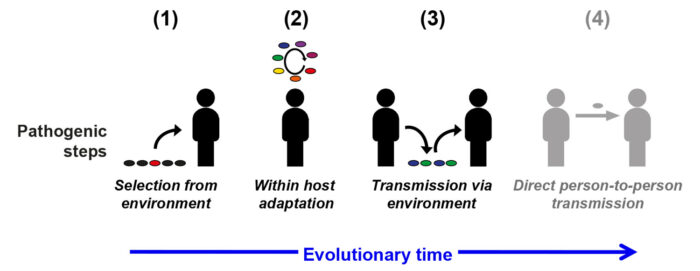
The results from this study point towards a generalisable model for mycobacterial pathogenic evolution. It also highlights the importance of key interventions, such as early treatment (to impede the bacterium’s ability to adapt within its host) and cross-infection control, which could prove vital in restricting the proliferation of existing pathogens such as M. abscessus, and prevent the emergence of new ones. Without this, it is likely that increased opportunities for direct transmission of pathogenic mycobacteria – because of factors such as rising population density or host susceptibility – will result in accelerated, uncontainable evolution into an obligate human pathogen, as occurred in M. tuberculosis several thousand years ago.
This work was funded by UKRI MRC, The Wellcome Trust, Botnar Foundation, The UK Cystic Fibrosis Trust, NIHR Cambridge Biomedical Research Centre.
Further References
Stepwise pathogenic evolution of Mycobacterium abscessus. Bryant, JM., Brown, KP., Burbaud, S., Everall, I., Belardinelli, JM., Rodriguez-Rincon, D., Grogono, DM., Peterson, CM., Verma, D., Evans, IE., Ruis, C., Weimann, A., Arora, D., Malhotra, S., Bannerman, B., Passemar, C., Templeton, K., MacGregor, G., Jiwa, K., Fisher, AJ., Blundell, TL., Ordway, DJ., Jackson, M., Parkhill, J., Floto, RA. Science, Vol. 372, Issue 6541.
Andres’ group page
Julian’s page