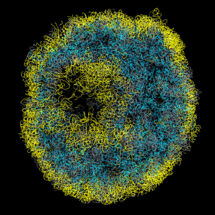
DNA in the nucleus is arranged into nucleosomes to produce an 11nm fibre which then intricately folds into high order assemblies. This nuclear organisation – the 3D arrangement of the genome within the nucleus – is critically linked to nuclear processes. Previously it has only been possible to analyse genome organisation across populations of cells. However, using a new chromosome conformation capture technique, Tim Stevens from the LMB’s Cell Biology Division, in collaboration with Ernest Laue’s group in the University of Cambridge Department of Biochemistry, has calculated the first 3D structures of entire mammalian genomes from single cells that have been selected using a microscope, allowing study of genome folding to a resolution of 100 kb.
David Lando and Srinjan Basu from Ernest’s group isolated and then chemically fixed single cells, to hold their DNA in place. The nuclei were extracted and many cuts made in the DNA strands. The cut ends of the DNA were then joined back together, but in a different way to the original arrangement, such that DNA strands in physical, but not necessarily linear, proximity were now connected. The DNA was then cut up even more and the fragments released. The small bits of DNA that had been linked together were selected and sequenced. By mapping these connections to the known sequence of the genome Tim determined which parts of the genome were close in space, uncovering the 3D arrangement of the chromosomes.
This revealed that whilst there are certain consistencies in nuclear organisation between cells, there is also substantial variation. Transcriptionally active and inactive regions segregate, with particularly active genes clustered, in a consistent way on a genome-wide basis in every cell. In contrast, the arrangement of DNA loops and domains vary markedly from cell to cell. These differences might help to explain how different cells, of notionally the same type, could behave differently as a result of the somewhat stochastic arrangement of chromosomes in their nuclei.
Whole genome structures, such as those pioneered in this study, are likely to become an important resource for developing and testing molecular hypotheses about how the different parts of a genome work together. Understanding how genes within the genome are organised in 3D will allow us to track the 3D changes that occur when cells develop into tissues or acquire diseases, providing clues about which genes are involved in these processes.
This work was funded by the MRC, Wellcome Trust and the European Commission.
Further references:
Article in Nature
Ernest Laue’s group page
MRC press release