The first structure of a eukaryotic SMC complex has been determined after two decades of intense study
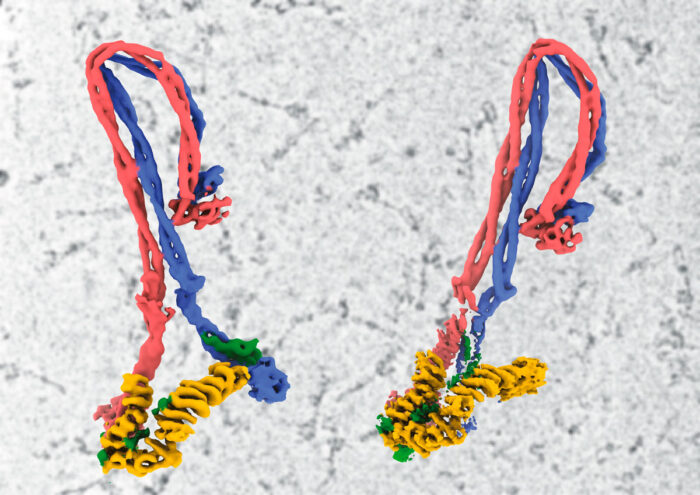
It is estimated that each of our cells contains about two metres of DNA. For our cells to be able to store all of this, the DNA must be very tightly compacted. This is achieved in a number of ways, involving many different proteins and complexes. One such complex known to have a role in compacting DNA into chromosomes is condensin, which loops DNA, however the mechanism by which it achieves this has taken decades to unravel. Now Jan Löwe’s group, in the LMB’s Structural Studies Division, in collaboration with researchers at EMBL and the MRC London Institute of Medical Sciences, have produced the first atomic model of condensin in its entirety, providing a clearer understanding of conformational changes during its reaction cycle.
Condensin is a large protein complex that plays a central role in chromosome assembly and segregation during mitosis and meiosis. Two of the core subunits of condensin are Smc2 and Smc4; two members of the SMC (structural maintenance of chromosomes) family of ATPases. Another important complex containing SMC proteins is cohesin, which, amongst other things, is responsible for holding sister chromatids together during the metaphase stage of mitosis. SMC complexes are ancient molecular machines found in all cells. Despite their fundamental roles, their precise molecular mechanisms have not yet been elucidated.
Although Jan’s group has been working on SMC complexes for about 20 years, it has been hard to even understand what they do with DNA in cells and determining the structures is made difficult by their highly elongated and floppy nature. Cryo-EM has now made this possible, with Byung-Gil Lee, a member of Jan’s group, leading the efforts to produce the first complete atomic model of a eukaryotic SMC complex. By capturing the machine in three different states, they have also been able to produce a model of how it might be enabled to move on DNA during its reaction cycle.
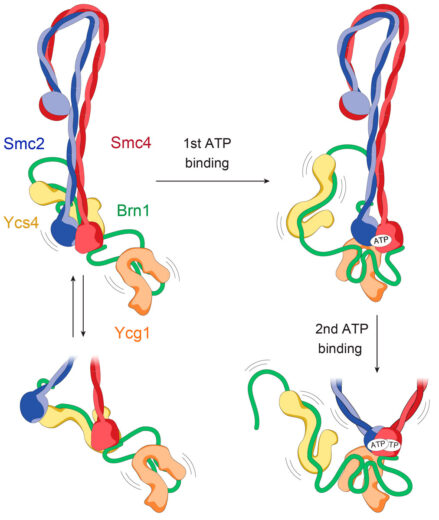
The core of condensin is made up of coiled coils of Smc2 and Smc4 in a highly unusual folded rod shape. Byung-Gil and the team have proposed a model in which binding of ATP at the “head” end leads to a more open conformation, which becomes the ring that encloses DNA, rather than directly binding to it.
This work lays the foundation for future work that will look at how these complexes interact with DNA in order to create loops of DNA and compact it into chromosomes. Condensin is required for cell cycle progression, a process that, when dysregulated, can lead to cancer, so an improved understanding of the functions and mechanisms of SMC complexes might have applications for cancer therapy.
The work was funded by UKRI MRC, EMBL, ERC, DFG, and Wellcome.
Further references
Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Lee, B-G., Merkel, F., Allegretti, M., Hassler, M., Cawood, C., Lecomte, L., O’Reilly, FJ., Sinn, LR., Gutierrez-Escribano, P., Kschonsak, M., Bravo, S., Nakane, T., Rappsilber, J., Aragon, L., Beck, M., Löwe, J., Haering, CH. Nature Structural and Molecular Biology (Epub ahead of print)
Jan’s group page
Luis Aragón’s group page
Martin Beck’s group page
Christian Häring’s group page