Previously unseen structural flexibility of N-terminal domains is crucial for regulating speed by which GluA1 opens and closes ion channel gate, and recruiting receptor into synapse
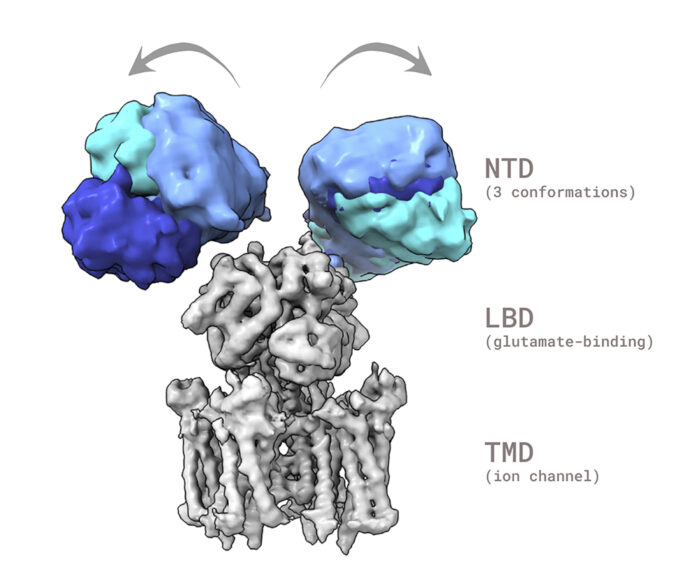
AMPA receptors (AMPARs) are glutamate-gated cation channels, composed of four core subunits – GluA1 to GluA4 – and are the primary mediators of excitatory neurotransmission throughout the brain, playing a central role in learning and memory at synapses. There are two primary types of receptors: GluA2-containing, Ca2+– impermeable, and GluA2-lacking, Ca2+-permeable (CP). All structural studies have focused on the former, which dominate in the brain. There is currently no structural knowledge concerning GluA2-lacking receptors, despite their prominent expression in glia and interneurons, and their involvement in long-term potentiation and a range of neuropathologies. Ingo Greger’s group, in the LMB’s Neurobiology Division, has now published the first structures of a GluA2-lacking AMPAR, the GluA1 AMPA receptor homomer, a prominent CP AMPAR.
Group members Josip Ivica and Imogen Stockwell functionally characterised AMPA receptors from mammalian cells, trapped in various functional states with specific ligands and at neuronal synapses. Danyang Zhang, another member of Ingo’s group, then used electron cryo-microscopy (cryo-EM) to determine the structures, in three different states of the gating spectrum. Structural analysis was supported by James Krieger at the National Centre for Biotechnology of the Spanish National Research Council (CNB-CSIC), Madrid, Spain.
This study revealed a previously unseen structural flexibility, which is particularly prominent in the sequence-diverse N-terminal domains (NTDs). This NTD mobility triggers rearrangement of the receptor subunits leading to slowed gating kinetics, and also impacts recruitment of the receptor into the synapse. The group further experimented by introducing an NTD point mutation into otherwise stable GluA2 receptors, resulting in similar structural dynamics to GluA1 and comparable functional consequences. This illustrates that NTDs work as a key switch to determine AMPAR gating kinetics and ultimately their signalling at the synapse.
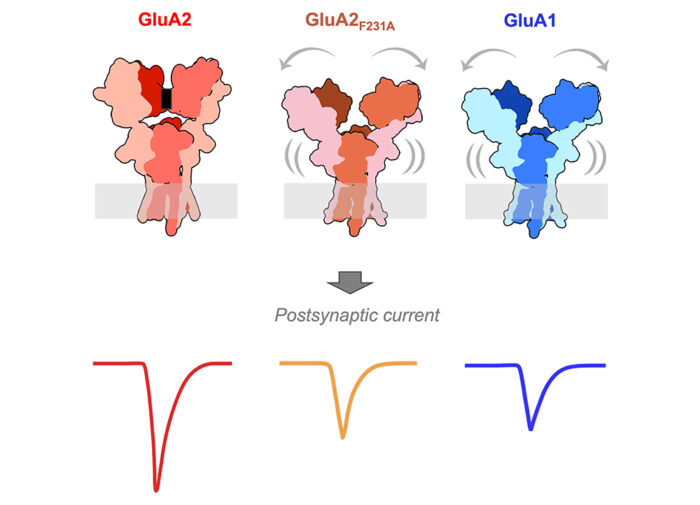
This research is significant as glutamate receptors are central to learning and memory at synapses in the brain. Memory formation involves the activation of a synapse, which involves the recruitment and accumulation of GluA1-type AMPA receptors. This study gives new insight into this process, notably the fact that NTDs are needed to regulate GluA1’s gating kinetics, and the way the receptor is recruited into the synapse. This opens new avenues of research to further investigate the operation and synaptic recruitment of GluA1 AMPA receptors during learning.
Additionally, the malfunction of glutamate receptors is associated with a variety of neurological and neuropsychiatric disorders, such as epilepsy, strokes, schizophrenia and depression. Further research on their mechanism of action could point to NTDs as a potential drug target for therapeutic interventions.
This work was funded by UKRI MRC and the Wellcome Trust.

In addition, this project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 748478 .
Further references
Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor. Zhang, D., Ivica, J., Krieger, JM., Ho, H., Yamashita, K., Stockwell, I., Baradaran, R., Cais, O., Greger, IH. Nature
Ingo’s group page
Previous Insight on Research articles
New structures show how auxiliary subunits modulate hippocampal AMPA receptor neurotransmission
Architecture of a prominent neurotransmitter receptor involved in memory formation and learning revealed