 The first X-ray crystal structure of the motor domain of cytoplasmic dynein, a protein that uses the cellular energy from ATP to walk along microtubule tracks that run throughout the cell, has been solved.
The first X-ray crystal structure of the motor domain of cytoplasmic dynein, a protein that uses the cellular energy from ATP to walk along microtubule tracks that run throughout the cell, has been solved.
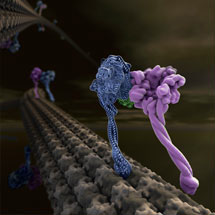
Cytoplasmic dynein moves numerous cargos around the cell including proteins and RNAs that set up the cell polarity, membraneous organelles, aggregated proteins that are toxic unless collected and disposed of, and even whole nuclei. Many viruses need to hijack dynein in order to get to the nucleus and infect the cell.
The crystal structure of dynein represents one of the most anticipated results in the motor protein/cytoskeleton field. Until now, the massive atomic size of this molecular machine has made it a challenging target for structural work and only low-resolution EM images of the motor protein were available.
The research was carried out over the last seven years by Andrew Carter, a new group leader in LMB’s Structural Studies Division, and Carol Cho, a graduate student who worked with Andrew in Ron Vale’s lab at UCSF, where the majority of the research was completed.
Andrew and Carol determined a low-resolution crystal structure that allowed them to build a complete model for the 300kD motor domain. Cytoplasmic dynein contains a dimer of two motor domains that take alternate steps to walk along the microtubule. As their crystal structure is also a dimer it gives some insight into how this walking can occur.
Dynein is involved with many diseases, from the transport of viruses, to developmental disorders such as lissencephaly, a neuronal migration defect. Though there are no immediate implications from knowing this structure to treating these disorders, an understanding of what goes wrong in lissencephaly will benefit from this work, as lissencephaly results from lack of a protein LIS1 that interacts with the dynein motor.
The current research in Andrew’s lab aims to get a higher resolution view of the motor domain. This will complete the story by allowing the position of all 2700 amino acids to be determined and reveal which of the four possible binding sites for ATP are occupied and in turn which ones are important for dynein’s walk.
The research was funded by NIH, HHMI, the Leukemia Lymphoma Society (USA), the Jane Coffin Childs Fund and the MRC.
Further references:
Full article in Science
Science commentary
Andrew Carter’s Group Page
C&EN news article: ‘Dynein’s Motor Revealed’