Manu Hegde’s group shows how ribosome collisions along an mRNA reduce additional ribosome traffic until the collisions are cleared
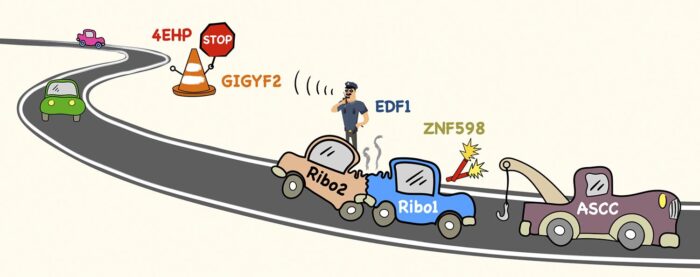
Cells contain several million ribosomes – macromolecular machines that interpret the genetic code in strands of messenger RNA (mRNA) to produce proteins. This process, called translation, involves individual ribosomes reading along the mRNA from beginning to end. To maximise efficiency, new ribosomes start translating an mRNA before previous ones have finished. Thus, most mRNAs in the cell have many ribosomes simultaneously translating along its length, like cars speeding along a busy road. Manu Hegde’s group in the LMB’s Cell Biology Division have now provided new insights into how ribosome collisions on a crowded mRNA are managed to avoid major ribosome traffic jams.
To understand how cells deal with ribosome collisions, Szymon Juszkiewicz from Manu’s lab used a ribosome inhibitor to cause widespread collisions inside cells. By comparing all the proteins associated with collided versus non-collided ribosomes, Szymon discovered two collision-specific factors called EDF1 and GIGYF2. He found that without either of these factors, mRNAs containing collisions are translated more often than they would be normally.
Earlier work by others had shown that GIGYF2 binds to a protein called 4EHP, which can block the front of an mRNA. Putting these pieces together, Szymon found that EDF1 detects collided ribosomes, recruits GIGYF2 and 4EHP, which blocks the mRNA to temporarily prevent new ribosomes from starting translation. This relay system is like a policeman calling to close road access until an accident has been cleared.
Although many collisions resolve quickly as ribosomes continue translation, a ribosome sometimes gets stuck (for example at a site of mRNA damage), causing trailing ribosomes to collide. Earlier work from Manu’s lab had shown that ribosome collisions are marked by an enzyme called ZNF598, similarly to how road flares are used to mark accident sites on a road. For ribosomes, a small protein called ubiquitin serves as the flare. What happens next was not clear, but genetic studies had implicated a factor called the ASC-1 complex (ASCC).
Through a chance meeting, Szymon learned that Shaun Speldewinde and Li Wan in Jesper Svejstrup’s group at the Francis Crick Institute had purified ASCC for another reason. Szymon showed that purified ASCC disassembles a stuck ribosome marked with ubiquitin to clear the way for ribosomes behind it to continue translating. Thus, Szymon’s two studies collectively show that a ribosome collision on the mRNA “road” not only regulates entering traffic, but also calls the equivalent of a tow truck to clear the wreckage.
The new research is important because the quality and amounts of proteins in a cell are crucial for that cell’s function. An excess of defective proteins, such as those produced by stuck ribosomes, can cause neurodegeneration. Similarly, mutations in GIGYF2 cause a Parkinson’s like neurodegeneration in humans through unknown mechanisms. Finally, alterations in protein translation rates is associated with many other diseases ranging from viral infection to cancer. Thus, understanding how cells fine tune translation to avoid ribosome traffic jams, and how serious errors are cleared, could provide new avenues for intervention in diseases where these pathways are defective.
The work was funded by UKRI MRC, CRUK, Wellcome Trust, and ERC.
Further references:
Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. Juszkiewicz, S., Slodkowicz, G., Lin, Z., Freire-Pritchett, P., Peak-Chew, SY., and Hegde, RS. eLife (Epub ahead of print)
The ASC-1 Complex Disassembles Collided Ribosomes. Juszkiewicz, S., Speldewinde, SH., Wan, L., Svejstrup, JQ., and Hegde, RS. Molecular Cell (Epub ahead of print)
Manu’s group page
Jesper Svejstrup’s group page